Inhibition of virus production in JC virus-infected cells by postinfection RNA interference
- PMID: 15194803
- PMCID: PMC421637
- DOI: 10.1128/JVI.78.13.7270-7273.2004
Inhibition of virus production in JC virus-infected cells by postinfection RNA interference
Abstract
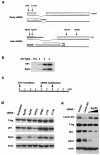
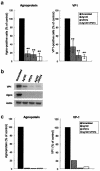
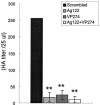
RNA interference has been applied for the prevention of virus infections in mammalian cells but has not succeeded in eliminating infections from already infected cells. We now show that the transfection of JC virus-infected SVG-A human glial cells with small interfering RNAs that target late viral proteins, including agnoprotein and VP1, results in a marked inhibition both of viral protein expression and of virus production. RNA interference directed against JC virus genes may thus provide a basis for the development of new strategies to control infections with this polyomavirus.
Figures
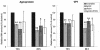
Similar articles
-
Intracellular approach for blocking JC virus gene expression by using RNA interference during viral infection.J Virol. 2004 Jul;78(13):7264-9. doi: 10.1128/JVI.78.13.7264-7269.2004. J Virol. 2004. PMID: 15194802 Free PMC article.
-
[Recent research on the JC virus].No To Shinkei. 2007 Feb;59(2):101-8. No To Shinkei. 2007. PMID: 17315751 Review. Japanese.
-
Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1.J Virol. 2002 Apr;76(8):3828-38. doi: 10.1128/jvi.76.8.3828-3838.2002. J Virol. 2002. PMID: 11907223 Free PMC article.
-
Role of JC virus agnoprotein in virion formation.Microbiol Immunol. 2012 Sep;56(9):639-46. doi: 10.1111/j.1348-0421.2012.00484.x. Microbiol Immunol. 2012. PMID: 22708997
-
[Recent research on the JC virus].Brain Nerve. 2007 Feb;59(2):101-8. Brain Nerve. 2007. PMID: 17380774 Review. Japanese.
Cited by
-
Progressive multifocal leukoencephalopathy: current treatment options and future perspectives.Ther Adv Neurol Disord. 2015 Nov;8(6):255-73. doi: 10.1177/1756285615602832. Ther Adv Neurol Disord. 2015. PMID: 26600871 Free PMC article. Review.
-
Progressive multifocal leukoencephalopathy: clinical and molecular aspects.Rev Med Virol. 2012 Jan;22(1):18-32. doi: 10.1002/rmv.710. Epub 2011 Sep 21. Rev Med Virol. 2012. PMID: 21936015 Free PMC article. Review.
-
Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus agnoprotein: role in nuclear egress of viral particles.EMBO Rep. 2005 May;6(5):452-7. doi: 10.1038/sj.embor.7400406. EMBO Rep. 2005. PMID: 15864296 Free PMC article.
-
Avian metapneumovirus phosphoprotein targeted RNA interference silences the expression of viral proteins and inhibits virus replication.Antiviral Res. 2006 Jan;69(1):46-51. doi: 10.1016/j.antiviral.2005.09.004. Epub 2005 Nov 15. Antiviral Res. 2006. PMID: 16310868 Free PMC article.
-
Regulation of gene expression in primate polyomaviruses.J Virol. 2009 Nov;83(21):10846-56. doi: 10.1128/JVI.00542-09. Epub 2009 Jul 29. J Virol. 2009. PMID: 19640999 Free PMC article. Review.
References
-
- Andino, R. 2003. RNAi puts a lid on virus replication. Nat. Biotechnol. 21:629-630. - PubMed
-
- Clifford, D. B. 1999. Opportunistic viral infections in the setting of human immunodeficiency virus. Semin. Neurol. 19:185-192. - PubMed
-
- Endo, S., Y. Okada, Y. Orba, H. Nishihara, S. Tanaka, K. Nagashima, and H. Sawa. 2003. JC virus agnoprotein colocalizes with tubulin. J. Neurovirol. 9(Suppl. 1):10-14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

