Structural requirements of the glucocorticoid-response unit of the carbamoyl-phosphate synthase gene
- PMID: 15196051
- PMCID: PMC1133802
- DOI: 10.1042/BJ20040471
Structural requirements of the glucocorticoid-response unit of the carbamoyl-phosphate synthase gene
Abstract
The GRU (glucocorticoid-response unit) within the distal enhancer of the gene encoding carbamoyl-phosphate synthase, which comprises REs (response elements) for the GR (glucocorticoid receptor) and the liver-enriched transcription factors FoxA (forkhead box A) and C/EBP (CCAAT/enhancer-binding protein), and a binding site for an unknown protein denoted P3, is one of the simplest GRUs described. In this study, we have established that the activity of this GRU depends strongly on the positioning and spacing of its REs. Mutation of the P3 site within the 25 bp FoxA-GR spacer eliminated GRU activity, but the requirement for P3 could be overcome by decreasing the length of this spacer to < or =12 bp, by optimizing the sequence of the REs in the GRU, and by replacing the P3 sequence with a C/EBPbeta sequence. With spacers of < or =12 bp, the activity of the GRU depended on the helical orientation of the FoxA and GR REs, with highest activities observed at 2 and 12 bp respectively. Elimination of the 6 bp C/EBP-FoxA spacer also increased GRU activity 2-fold. Together, these results indicate that the spatial positioning of the transcription factors that bind to the GRU determines its activity and that the P3 complex, which binds to the DNA via a 75 kDa protein, functions to facilitate interaction between the FoxA and glucocorticoid response elements when the distance between these transcription factors means that they have difficulties contacting each other.
Figures

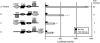






References
-
- Meijer A. J., Lamers W. H., Chamuleau R. A. Nitrogen metabolism and ornithine cycle function. Physiol. Rev. 1990;70:701–748. - PubMed
-
- Christoffels V. M., van den Hoff M. J., Moorman A. F., Lamers W. H. The far-upstream enhancer of the carbamoyl-phosphate synthetase I gene is responsible for the tissue specificity and hormone inducibility of its expression. J. Biol. Chem. 1995;270:24932–24940. - PubMed
-
- Christoffels V. M., Grange T., Kaestner K. H., Cole T. J., Darlington G. J., Croniger C. M., Lamers W. H. Glucocorticoid receptor, C/EBP, HNF3, and protein kinase A coordinately activate the glucocorticoid response unit of the carbamoylphosphate synthetase I gene. Mol. Cell. Biol. 1998;18:6305–6315. - PMC - PubMed
-
- Mitchell J., Noisin E., Hall R., O'Brien R., Imai E., Granner D. Integration of multiple signals through a complex hormone response unit in the phosphoenolpyruvate carboxykinase gene promoter. Mol. Endocrinol. 1994;8:585–594. - PubMed
-
- Wedel A., Ziegler-Heitbrock H. W. The C/EBP family of transcription factors. Immunobiology. 1995;193:171–185. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

