Comparative sequence and x-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse
- PMID: 15197169
- PMCID: PMC442142
- DOI: 10.1101/gr.2575904
Comparative sequence and x-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse
Abstract
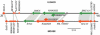
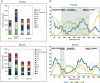
We have performed X-inactivation and sequence analyses on 350 kb of sequence from human Xp11.2, a region shown previously to contain a cluster of genes that escape X inactivation, and we compared this region with the region of conserved synteny in mouse. We identified several new transcripts from this region in human and in mouse, which defined the full extent of the domain escaping X inactivation in both species. In human, escape from X inactivation involves an uninterrupted 235-kb domain of multiple genes. Despite highly conserved gene content and order between the two species, Smcx is the only mouse gene from the conserved segment that escapes inactivation. As repetitive sequences are believed to facilitate spreading of X inactivation along the chromosome, we compared the repetitive sequence composition of this region between the two species. We found that long terminal repeats (LTRs) were decreased in the human domain of escape, but not in the majority of the conserved mouse region adjacent to Smcx in which genes were subject to X inactivation, suggesting that these repeats might be excluded from escape domains to prevent spreading of silencing. Our findings indicate that genomic context, as well as gene-specific regulatory elements, interact to determine expression of a gene from the inactive X-chromosome.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
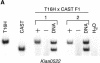
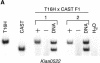


Similar articles
-
Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine.Genome Res. 2002 Jun;12(6):894-908. doi: 10.1101/gr.152902. Genome Res. 2002. PMID: 12045143 Free PMC article.
-
Chromosomal domains and escape from X inactivation: comparative X inactivation analysis in mouse and human.Mamm Genome. 2000 Oct;11(10):849-54. doi: 10.1007/s003350010175. Mamm Genome. 2000. PMID: 11003698
-
The Drosophila developmental gene fat facets has a human homologue in Xp11.4 which escapes X-inactivation and has related sequences on Yq11.2.Hum Mol Genet. 1996 Nov;5(11):1695-701. doi: 10.1093/hmg/5.11.1695. Hum Mol Genet. 1996. PMID: 8922996
-
A stain upon the silence: genes escaping X inactivation.Trends Genet. 2003 Aug;19(8):432-8. doi: 10.1016/S0168-9525(03)00177-X. Trends Genet. 2003. PMID: 12902161 Review.
-
Escape from X inactivation.Cytogenet Genome Res. 2002;99(1-4):36-43. doi: 10.1159/000071572. Cytogenet Genome Res. 2002. PMID: 12900543 Review.
Cited by
-
Mutations in epilepsy and intellectual disability genes in patients with features of Rett syndrome.Am J Med Genet A. 2015 Sep;167A(9):2017-25. doi: 10.1002/ajmg.a.37132. Epub 2015 Apr 25. Am J Med Genet A. 2015. PMID: 25914188 Free PMC article.
-
X chromosome choice occurs independently of asynchronous replication timing.J Cell Biol. 2005 Jan 31;168(3):365-73. doi: 10.1083/jcb.200405117. Epub 2005 Jan 24. J Cell Biol. 2005. PMID: 15668296 Free PMC article.
-
Human Coronavirus Infection Reorganizes Spatial Genomic Architecture in Permissive Lung Cells.Res Sq [Preprint]. 2024 Mar 13:rs.3.rs-3979539. doi: 10.21203/rs.3.rs-3979539/v1. Res Sq. 2024. PMID: 38559036 Free PMC article. Preprint.
-
Epigenetic predisposition to expression of TIMP1 from the human inactive X chromosome.BMC Genet. 2005 Sep 29;6:48. doi: 10.1186/1471-2156-6-48. BMC Genet. 2005. PMID: 16194278 Free PMC article.
-
Dosage compensation and gene expression on the mammalian X chromosome: one plus one does not always equal two.Chromosome Res. 2009;17(5):637-48. doi: 10.1007/s10577-009-9063-9. Chromosome Res. 2009. PMID: 19802704 Free PMC article. Review.
References
-
- Agulnik, A.I., Mitchell, M.J., Lerner, J.L., Woods, D.R., and Bishop, C.E. 1994a. A mouse Y chromosome gene encoded by a region essential for spermatogenesis and expression of male-specific minor histocompatibility antigens. Hum. Mol. Genet. 3: 873–878. - PubMed
-
- Agulnik, A.I., Mitchell, M.J., Mattei, M.G., Borsani, G., Avner, P.A., Lerner, J.L., and Bishop, C.E. 1994b. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum. Mol. Genet. 3: 879–884. - PubMed
-
- Agulnik, A.I., Longepied, G., Ty, M.T., Bishop, C.E., and Mitchell, M. 1999. Mouse H-Y encoding Smcy gene and its X chromosomal homolog Smcx. Mamm. Genome 10: 926–929. - PubMed
WEB SITE REFERENCES
-
- http://bio.cse.psu.edu/pipmaker/; PipMaker and MultiPipMaker.
-
- http://bioinformatics.weizmann.ac.il/cards/; GeneCards.
-
- http://ftp.genome.washington.edu/; RepeatMasker.
-
- http://genome.ucsc.edu/; USCS Genome Biotechnology.
-
- http://grail.lsd.ornl.gov/grailexp/; Computational Biology at ORNL.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
