Protein tolerance to random amino acid change
- PMID: 15197260
- PMCID: PMC438954
- DOI: 10.1073/pnas.0403255101
Protein tolerance to random amino acid change
Abstract
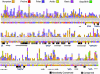
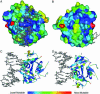
Mutagenesis of protein-encoding sequences occurs ubiquitously; it enables evolution, accumulates during aging, and is associated with disease. Many biotechnological methods exploit random mutations to evolve novel proteins. To quantitate protein tolerance to random change, it is vital to understand the probability that a random amino acid replacement will lead to a protein's functional inactivation. We define this probability as the "x factor." Here, we develop a broadly applicable approach to calculate x factors and demonstrate this method using the human DNA repair enzyme 3-methyladenine DNA glycosylase (AAG). Three gene-wide mutagenesis libraries were created, each with 10(5) diversity and averaging 2.2, 4.6, and 6.2 random amino acid changes per mutant. After determining the percentage of functional mutants in each library using high-stringency selection (>19,000-fold), the x factor was found to be 34% +/- 6%. Remarkably, reanalysis of data from studies of diverse proteins reveals similar inactivation probabilities. To delineate the nature of tolerated amino acid substitutions, we sequenced 244 surviving AAG mutants. The 920 tolerated substitutions were characterized by substitutability index and mapped onto the AAG primary, secondary, and known tertiary structures. Evolutionarily conserved residues show low substitutability indices. In AAG, beta strands are on average less substitutable than alpha helices; and surface loops that are not involved in DNA binding are the most substitutable. Our results are relevant to such diverse topics as applied molecular evolution, the rate of introduction of deleterious alleles into genomes in evolutionary history, and organisms' tolerance of mutational burden.
Figures
References
-
- Harris, R. S., Sheehy, A. M., Craig, H. M., Malim, M. H. & Neuberger, M. S. (2003) Nat. Immunol. 4, 641-643. - PubMed
-
- Smith, J. M. (1970) Nature 225, 563-564. - PubMed
-
- Creighton, T. E. (1993) Proteins (Freeman, New York).
-
- Bowie, J. U., Reidhaar-Olson, J. F., Lim, W. A. & Sauer, R. T. (1990) Science 247, 1306-1310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

