High-resolution molecular characterization of 15q11-q13 rearrangements by array comparative genomic hybridization (array CGH) with detection of gene dosage
- PMID: 15197683
- PMCID: PMC1216061
- DOI: 10.1086/422854
High-resolution molecular characterization of 15q11-q13 rearrangements by array comparative genomic hybridization (array CGH) with detection of gene dosage
Abstract
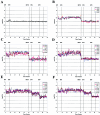
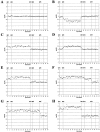
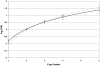
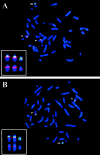
Maternally derived duplication of the imprinted region of chromosome 15q11-q14 leads to a complex neurobehavioral phenotype that often includes autism, cognitive deficits, and seizures. Multiple repeat elements within the region mediate a variety of rearrangements, including interstitial duplications, interstitial triplications, and supernumerary isodicentric marker chromosomes, as well as the deletions that cause Prader-Willi and Angelman syndromes. To elucidate the molecular structure of these duplication chromosomes, we designed a high-resolution array comparative genomic hybridization (array CGH) platform. The array contains 79 clones that form a gapped contig across the critical region on chromosome 15q11-q14 and 21 control clones from other autosomes and the sex chromosomes. We used this array to examine a set of 48 samples from patients with segmental aneuploidy of chromosome 15q. Using the array, we were able to determine accurately the dosage, which ranged from 1 to 6 copies, and also to detect atypical and asymmetric rearrangements. In addition, the increased resolution of the array allowed us to position two previously reported breakpoints within the contig. These results indicate that array CGH is a powerful technique to study rearrangements of proximal chromosome 15q.
Figures


References
Electronic-Database Information
-
- BAC/PAC Resources Center, http://bacpac.chori.org (for BAC and PAC clones)
-
- Genome Database, http://www.gdb.org (for microsatellite markers)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
-
- UCSC Genome Browser, http://genome.ucsc.edu (for clone positions)
References
-
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

