Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression
- PMID: 15198975
- PMCID: PMC443518
- DOI: 10.1101/gad.299904
Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression
Abstract
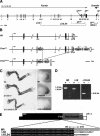
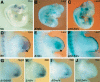
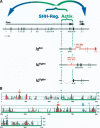
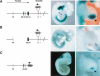
The mouse limb deformity (ld) mutations cause limb malformations by disrupting epithelial-mesenchymal signaling between the polarizing region and the apical ectodermal ridge. Formin was proposed as the relevant gene because three of the five ld alleles disrupt its C-terminal domain. In contrast, our studies establish that the two other ld alleles directly disrupt the neighboring Gremlin gene, corroborating the requirement of this BMP antagonist for limb morphogenesis. Further doubts concerning an involvement of Formin in the ld limb phenotype are cast, as a targeted mutation removing the C-terminal Formin domain by frame shift does not affect embryogenesis. In contrast, the deletion of the corresponding genomic region reproduces the ld limb phenotype and is allelic to mutations in Gremlin. We resolve these conflicting results by identifying a cis-regulatory region within the deletion that is required for Gremlin activation in the limb bud mesenchyme. This distant cis-regulatory region within Formin is also altered by three of the ld mutations. Therefore, the ld limb bud patterning defects are not caused by disruption of Formin, but by alteration of a global control region (GCR) required for Gremlin transcription. Our studies reveal the large genomic landscape harboring this GCR, which is required for tissue-specific coexpression of two structurally and functionally unrelated genes.
Figures



References
-
- Avsian-Kretchmer O. and Hsueh, A.J. 2003. Comparative genomic analysis of the eight-membered-ring cystine-knot-containing bone morphogenetic protein (BMP) antagonists. Mol. Endocrinol. 18: 1-12. - PubMed
-
- Bacchelli C., Goodman, F.R., Scambler, P.J., and Winter, R.M. 2001. Cenani-Lenz syndrome with renal hypoplasia is not linked to FORMIN or GREMLIN. Clin. Genet. 59: 203-205. - PubMed
-
- Evangelista M., Zigmond, S., and Boone, C. 2003. Formins: Signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116: 2603-2611. - PubMed
-
- Faustino N.A. and Cooper, T.A. 2003. Pre-mRNA splicing and human disease. Genes & Dev. 17: 419-437. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
