The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex
- PMID: 15198981
- PMCID: PMC423190
- DOI: 10.1101/gad.1199404
The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex
Abstract
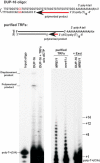
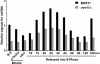
The precise DNA arrangement at chromosomal ends and the proteins involved in its maintenance are of crucial importance for genome stability. For the yeast Saccharomyces cerevisiae, this constitutive DNA configuration has remained unknown. We demonstrate here that G-tails of 12-14 bases are present outside of S phase on normal yeast telomeres. Furthermore, the Mre11p protein is essential for the proper establishment of this constitutive end-structure. However, the timing of extended G-tails occurring during S phase is not affected in strains lacking Mre11p. Thus, G-tails are present on yeast chromosomes throughout the cell cycle and the MRX complex is required for their normal establishment.
Figures



References
-
- Blackburn E.H. 2001. Switching and signaling at the telomere. Cell 106: 661-673. - PubMed
-
- Chakhparonian M. and Wellinger, R.J. 2003. Telomere maintenance and DNA replication: How closely are these two connected? Trends Genet. 19: 439-446. - PubMed
-
- ____. 2002. The Mre11 complex: At the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3: 317-327. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
