Synergistic transcription activation by Maf and Sox and their subnuclear localization are disrupted by a mutation in Maf that causes cataract
- PMID: 15199128
- PMCID: PMC480896
- DOI: 10.1128/MCB.24.13.5694-5709.2004
Synergistic transcription activation by Maf and Sox and their subnuclear localization are disrupted by a mutation in Maf that causes cataract
Abstract
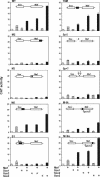
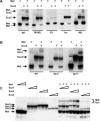
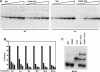
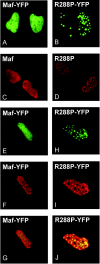
Crystallin genes are selectively expressed during lens development. Maf and Sox family proteins synergistically enhanced gammaF-crystallin promoter activity in a lens cell line. Mutational analysis of the gammaF-crystallin promoter identified a composite regulatory element containing nonconsensus Maf and Sox recognition sequences. Mutations in these recognition sequences or changes in their spacing eliminated synergistic transcription activation. The transcriptional synergy was also affected by changes in the orientation of the Maf recognition sequence that had no detectable effect on binding affinity. The interaction between Maf and Sox proteins was visualized in living cells by bimolecular fluorescence complementation analysis. The N-terminal region of Maf mediated the interaction with Sox proteins in cells. Synergistic transcription activation required the N-terminal region of Maf as well as the ancillary DNA binding domain and the unique portion of the basic region that mediate specific recognition of the gammaF-crystallin promoter element. A mutation in the ancillary DNA binding domain of Maf (R288P) that has been shown to cause cataract eliminated the transcriptional activity of Maf but had no detectable effect on DNA binding in vitro. Whereas wild-type Maf was uniformly distributed in the nucleoplasm, R288P Maf was enriched in nuclear foci. Cajal bodies and gemini of coiled bodies were closely associated with the foci occupied by R288P Maf. Wild-type Maf formed complexes with Sox proteins in the nucleoplasm, whereas R288P Maf recruited Sox proteins as well as other interaction partners to the nuclear foci. The mislocalization of normal cellular proteins to these foci provides a potential explanation for the dominant disease phenotype of the R288P mutation in Maf.
Figures






Similar articles
-
CREB-binding protein/p300 co-activation of crystallin gene expression.J Biol Chem. 2002 Jul 5;277(27):24081-9. doi: 10.1074/jbc.M201821200. Epub 2002 Apr 9. J Biol Chem. 2002. PMID: 11943779
-
Analysis of non-crystallin lens fiber cell gene expression in c-Maf -/- mice.Mol Vis. 2003 Jun 30;9:288-94. Mol Vis. 2003. PMID: 12835653
-
c-Maf negatively regulates ARE-mediated detoxifying enzyme genes expression and anti-oxidant induction.Oncogene. 2002 Aug 8;21(34):5301-12. doi: 10.1038/sj.onc.1205642. Oncogene. 2002. PMID: 12149651
-
Roles of Maf family proteins in lens development.Dev Dyn. 2004 Mar;229(3):440-8. doi: 10.1002/dvdy.10467. Dev Dyn. 2004. PMID: 14991699 Review.
-
Congenital hereditary cataracts.Int J Dev Biol. 2004;48(8-9):1031-44. doi: 10.1387/ijdb.041854jg. Int J Dev Biol. 2004. PMID: 15558493 Review.
Cited by
-
Lens fiber cell differentiation and denucleation are disrupted through expression of the N-terminal nuclear receptor box of NCOA6 and result in p53-dependent and p53-independent apoptosis.Mol Biol Cell. 2010 Jul 15;21(14):2453-68. doi: 10.1091/mbc.e09-12-1031. Epub 2010 May 19. Mol Biol Cell. 2010. PMID: 20484573 Free PMC article.
-
Generation of Lens Progenitor Cells and Lentoid Bodies from Pluripotent Stem Cells: Novel Tools for Human Lens Development and Ocular Disease Etiology.Cells. 2022 Nov 6;11(21):3516. doi: 10.3390/cells11213516. Cells. 2022. PMID: 36359912 Free PMC article. Review.
-
Sox5 and c-Maf cooperatively induce Th17 cell differentiation via RORγt induction as downstream targets of Stat3.J Exp Med. 2014 Aug 25;211(9):1857-74. doi: 10.1084/jem.20130791. Epub 2014 Jul 29. J Exp Med. 2014. PMID: 25073789 Free PMC article.
-
Mutation update of transcription factor genes FOXE3, HSF4, MAF, and PITX3 causing cataracts and other developmental ocular defects.Hum Mutat. 2018 Apr;39(4):471-494. doi: 10.1002/humu.23395. Epub 2018 Jan 16. Hum Mutat. 2018. PMID: 29314435 Free PMC article. Review.
-
Signaling and Gene Regulatory Networks in Mammalian Lens Development.Trends Genet. 2017 Oct;33(10):677-702. doi: 10.1016/j.tig.2017.08.001. Epub 2017 Aug 31. Trends Genet. 2017. PMID: 28867048 Free PMC article. Review.
References
-
- Ambrosetti, D. C., H. R. Scholer, L. Dailey, and C. Basilico. 2000. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J. Biol. Chem. 275:23387-23397. - PubMed
-
- Brady, J. P., D. Garland, Y. Duglas-Tabor, W. G. Robison, Jr., A. Groome, and E. F. Wawrousek. 1997. Targeted disruption of the mouse alpha A-crystallin gene induces cataract and cytoplasmic inclusion bodies containing the small heat shock protein alpha B-crystallin. Proc. Natl. Acad. Sci. USA 94:884-889. - PMC - PubMed
-
- Chen, Q., D. H. Dowhan, D. Liang, D. D. Moore, and P. A. Overbeek. 2002. CREB-binding protein/p300 co-activation of crystallin gene expression. J. Biol. Chem. 277:24081-24089. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
