Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity
- PMID: 15199139
- PMCID: PMC480894
- DOI: 10.1128/MCB.24.13.5835-5843.2004
Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity
Erratum in
-
Correction for Danovi et al., "Amplification of Mdmx (or Mdm4) Directly Contributes to Tumor Formation by Inhibiting p53 Tumor Suppressor Activity".Mol Cell Biol. 2019 Jul 16;39(15):e00150-19. doi: 10.1128/MCB.00150-19. Print 2019 Aug 1. Mol Cell Biol. 2019. PMID: 31311845 Free PMC article. No abstract available.
Abstract
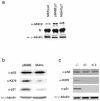
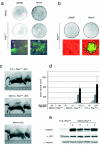
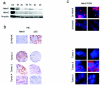
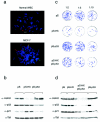
Human tumors are believed to harbor a disabled p53 tumor suppressor pathway, either through direct mutation of the p53 gene or through aberrant expression of proteins acting in the p53 pathway, such as p14(ARF) or Mdm2. A role for Mdmx (or Mdm4) as a key negative regulator of p53 function in vivo has been established. However, a direct contribution of Mdmx to tumor formation remains to be demonstrated. Here we show that retrovirus-mediated Mdmx overexpression allows primary mouse embryonic fibroblast immortalization and leads to neoplastic transformation in combination with HRas(V12). Furthermore, the human Mdmx ortholog, Hdmx, was found to be overexpressed in a significant percentage of various human tumors and amplified in 5% of primary breast tumors, all of which retained wild-type p53. Hdmx was also amplified and highly expressed in MCF-7, a breast cancer cell line harboring wild-type p53, and interfering RNA-mediated reduction of Hdmx markedly inhibited the growth potential of these cells in a p53-dependent manner. Together, these results make Hdmx a new putative drug target for cancer therapy.
Figures


References
-
- Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. - PubMed
-
- Bottger, V., A., Bottger, C., Garcia-Echeverria, Y. F., Ramos, A. J. van der Eb, A. G. Jochemsen, and D. P. Lane. 1999. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene 18:189-199. - PubMed
-
- Boyd, S. D., K. Y. Tsai, and T. Jacks. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563-568. - PubMed
-
- Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. - PubMed
-
- Colombo, E., J. C. Marine, D. Danovi, B. Falini, and P. G. Pelicci. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4:529-533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
