Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK
- PMID: 15199154
- PMCID: PMC480878
- DOI: 10.1128/MCB.24.13.6011-6020.2004
Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK
Abstract
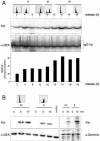
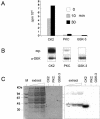
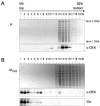
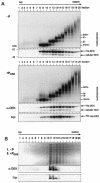
We have examined the posttranslational modification of the human chromatin protein DEK and found that DEK is phosphorylated by the protein kinase CK2 in vitro and in vivo. Phosphorylation sites were mapped by quadrupole ion trap mass spectrometry and found to be clustered in the C-terminal region of the DEK protein. Phosphorylation fluctuates during the cell cycle with a moderate peak during G(1) phase. Filter binding assays, as well as Southwestern analysis, demonstrate that phosphorylation weakens the binding of DEK to DNA. In vivo, however, phosphorylated DEK remains on chromatin. We present evidence that phosphorylated DEK is tethered to chromatin throughout the cell cycle by the un- or underphosphorylated form of DEK.
Figures




Similar articles
-
Functional domains of the ubiquitous chromatin protein DEK.Mol Cell Biol. 2004 Jul;24(13):6000-10. doi: 10.1128/MCB.24.13.6000-6010.2004. Mol Cell Biol. 2004. PMID: 15199153 Free PMC article.
-
The distribution of the DEK protein in mammalian chromatin.Biochem Biophys Res Commun. 2007 Jul 13;358(4):1008-14. doi: 10.1016/j.bbrc.2007.05.019. Epub 2007 May 21. Biochem Biophys Res Commun. 2007. PMID: 17524367
-
Phosphorylation of the oncogenic transcription factor interferon regulatory factor 2 (IRF2) in vitro and in vivo.J Cell Biochem. 1997 Aug 1;66(2):175-83. J Cell Biochem. 1997. PMID: 9213219
-
The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin.Gene. 2004 Dec 8;343(1):1-9. doi: 10.1016/j.gene.2004.08.029. Gene. 2004. PMID: 15563827 Review.
-
Progress in studies on the DEK protein and its involvement in cellular apoptosis.Sci China C Life Sci. 2009 Jul;52(7):637-42. doi: 10.1007/s11427-009-0088-2. Epub 2009 Jul 30. Sci China C Life Sci. 2009. PMID: 19641868 Review.
Cited by
-
Novel molecular mechanisms in Alzheimer's disease: The potential role of DEK in disease pathogenesis.Front Aging Neurosci. 2022 Oct 6;14:1018180. doi: 10.3389/fnagi.2022.1018180. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36275000 Free PMC article. Review.
-
Serum from mice immunized in the context of Treg inhibition identifies DEK as a neuroblastoma tumor antigen.BMC Immunol. 2007 Mar 30;8:4. doi: 10.1186/1471-2172-8-4. BMC Immunol. 2007. PMID: 17397536 Free PMC article.
-
Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions.Mol Cell Biol. 2006 Oct;26(20):7506-19. doi: 10.1128/MCB.00430-06. Epub 2006 Aug 7. Mol Cell Biol. 2006. PMID: 16894028 Free PMC article.
-
Fly Fishing for Histones: Catch and Release by Histone Chaperone Intrinsically Disordered Regions and Acidic Stretches.J Mol Biol. 2017 Aug 4;429(16):2401-2426. doi: 10.1016/j.jmb.2017.06.005. Epub 2017 Jun 10. J Mol Biol. 2017. PMID: 28610839 Free PMC article. Review.
-
High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length.Biochemistry. 2010 Aug 24;49(33):7119-30. doi: 10.1021/bi1004365. Biochemistry. 2010. PMID: 20669926 Free PMC article.
References
-
- Agresti, A., and M. E. Bianchi. 2003. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13:170-178. - PubMed
-
- Ahmed, K., D. A. Gerber, and C. Cochet. 2002. Joining the cell survival squad: an emerging role for protein kinase CK2. Trends Cell Biol. 12:226-230. - PubMed
-
- Allende, J. E., and C. C. Allende. 1995. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9:313-323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
