Structure-specific DNA binding and bipolar helicase activities of PcrA
- PMID: 15199167
- PMCID: PMC434446
- DOI: 10.1093/nar/gkh641
Structure-specific DNA binding and bipolar helicase activities of PcrA
Abstract
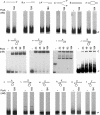
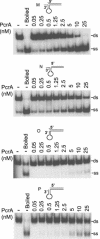
PcrA is an essential helicase in Gram-positive bacteria, but its precise role in cellular DNA metabolism is currently unknown. The Staphylococcus aureus PcrA helicase has both 5'-->3' and 3'-->5' helicase activities. In this work, we have studied the binding of S.aureus PcrA to a variety of DNA substrates that represent intermediates in DNA replication, repair, recombination and transcription. PcrA bound poorly or not at all to single-stranded DNA, double-stranded DNA with blunt ends, partially double-stranded DNA containing fork and bubble structures, and duplex DNA substrates containing either 5' or 3' single-stranded oligo dT tails. Interestingly, PcrA bound with high affinity to partially duplex DNA containing hairpin structures adjacent to a 6 nt long 5' single-stranded region and one unpaired nucleotide (flap) at the 3' end. However, PcrA did not detectably bind to partial duplexes with folded regions adjacent to a 6 nt long 3' single-stranded tail (with or without a 1 nt flap at the 5' end). PcrA showed moderate helicase activity with partially double-stranded DNAs containing 3' or 5' single-stranded oligo dT tails, the 3'-->5' helicase activity being more efficient than its 5'-->3' helicase activity. Interestingly, PcrA showed maximal helicase activity with substrates containing folded structures and 5' single-stranded tails, suggesting that its 5'-->3' helicase activity is greatly stimulated in the presence of specific structures. However, the 3'-->5' helicase activity of PcrA did not appear to be affected by the presence of folded substrates containing 3' single-stranded tails. Our data indicate that PcrA may recognize DNA substrates with specific structures in vivo and its 5'-->3' and 3'-->5' helicase activities may be involved in distinct cellular processes.
Figures


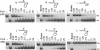
References
-
- Hall M.C. and Matson,S.W. (1999) Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol., 34, 867–877. - PubMed
-
- Bird L.E., Subramanya,S. and Wigley,D.B. (1998) Helicases: a unifying structural theme? Curr. Opin. Struct. Biol., 8, 14–18. - PubMed
-
- Marians K.J. (2000) Crawling and wiggling on DNA: structural insights to the mechanism of DNA unwinding by helicases. Structure, 8, R227–R235. - PubMed
-
- Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. - PubMed
-
- Iordanescu S. (1993) Identification and characterization of Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol. Gen. Genet., 241, 185–192. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

