Influenza A virus PB1-F2 gene in recent Taiwanese isolates
- PMID: 15200852
- PMCID: PMC3323094
- DOI: 10.3201/eid1004.030412
Influenza A virus PB1-F2 gene in recent Taiwanese isolates
Abstract
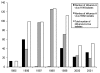
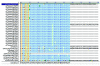
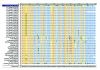
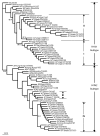
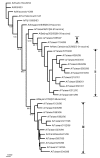
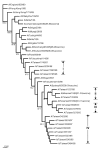
Influenza A virus contains eight RNA segments and encodes 10 viral proteins. However, an 11th protein, called PB1-F2, was found in A/Puerto Rico/8/34 (H1N1). This novel protein is translated from an alternative open reading frame (ORF) in the PB1 gene. We analyzed the PB1 gene of 42 recent influenza A isolates in Taiwan, including 24 H1N1 and 18 H3N2 strains. One H1N1 isolate and 17 H3N2 isolates contained the entire PB1-F2 ORF of 90 residues, three amino acids (aa) longer than the PB1-F2 of A/Puerto Rico/8/34 at the C terminal. The one remaining H3N2 isolate encoded a truncated PB1-F2 with 79 residues. The other 23 H1N1 isolates contained a truncated PB1-F2 of 57 aa. Phylogenetic analysis of both the HA and the PB1 genes showed that they shared similar clustering of these Taiwanese isolates, suggesting that no obvious reassortment occurred between the two genomic segments.
Figures
Similar articles
-
PB1-F2 gene in influenza a viruses of different hemagglutinin subtype.Acta Virol. 2006;50(4):269-72. Acta Virol. 2006. PMID: 17177613
-
Influenza a virus PB1-F2 protein.Acta Virol. 2007;51(2):101-8. Acta Virol. 2007. PMID: 17900216 Review.
-
Surveillance in Eastern India (2007-2009) revealed reassortment event involving NS and PB1-F2 gene segments among co-circulating influenza A subtypes.Virol J. 2012 Jan 5;9:3. doi: 10.1186/1743-422X-9-3. Virol J. 2012. PMID: 22217077 Free PMC article.
-
Prevalence of PB1-F2 of influenza A viruses.J Gen Virol. 2007 Feb;88(Pt 2):536-546. doi: 10.1099/vir.0.82378-0. J Gen Virol. 2007. PMID: 17251572
-
Current knowledge on PB1-F2 of influenza A viruses.Med Microbiol Immunol. 2011 May;200(2):69-75. doi: 10.1007/s00430-010-0176-8. Epub 2010 Oct 16. Med Microbiol Immunol. 2011. PMID: 20953627 Review.
Cited by
-
Differential localization and function of PB1-F2 derived from different strains of influenza A virus.J Virol. 2010 Oct;84(19):10051-62. doi: 10.1128/JVI.00592-10. Epub 2010 Jul 21. J Virol. 2010. PMID: 20660199 Free PMC article.
-
Genomic signatures of human versus avian influenza A viruses.Emerg Infect Dis. 2006 Sep;12(9):1353-60. doi: 10.3201/eid1209.060276. Emerg Infect Dis. 2006. PMID: 17073083 Free PMC article.
-
Computational analysis and mapping of novel open reading frames in influenza A viruses.PLoS One. 2014 Dec 15;9(12):e115016. doi: 10.1371/journal.pone.0115016. eCollection 2014. PLoS One. 2014. PMID: 25506939 Free PMC article.
-
A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA.J Virol. 2009 Aug;83(16):8021-31. doi: 10.1128/JVI.00826-09. Epub 2009 Jun 3. J Virol. 2009. PMID: 19494001 Free PMC article.
-
Influenza A virus PB1-F2 gene.Emerg Infect Dis. 2006 Oct;12(10):1607-8; author reply 1608-9. doi: 10.3201/eid1210.060511. Emerg Infect Dis. 2006. PMID: 17176587 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
