Exchange protein directly activated by cAMP (EPAC) interacts with the light chain (LC) 2 of MAP1A
- PMID: 15202935
- PMCID: PMC1133955
- DOI: 10.1042/BJ20040122
Exchange protein directly activated by cAMP (EPAC) interacts with the light chain (LC) 2 of MAP1A
Abstract
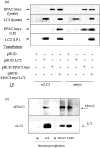
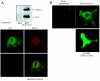
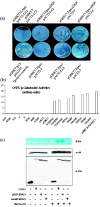
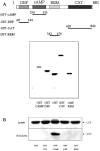
Using EPAC1 (exchange protein directly activated by cAMP 1) as bait in two-hybrid screens of foetal and adult human brain libraries, we identified the LC2 (light chain 2) of MAP1A (microtubule-associated protein 1A) as a protein capable of interaction with EPAC1. We applied an immunoprecipitation assay to demonstrate protein interaction between EPAC1 and LC2 in co-transfected human embryonic kidney 293 cells. EPAC2 also co-immunoprecipitated with LC2 from extracts of rat cerebellum. Immunolocalization in co-transfected human embryonic kidney 293 cells revealed that EPAC1 co-localizes with LC2 throughout the cell body. We found that endogenous EPAC2 is also immunolocalized with LC2 in PC12 cells. Immunolocalization of EPAC1 in transfected COS1 cells showed that EPAC1 is associated with the perinuclear region surrounding the nucleus and filamentous structures throughout the cell. Removal of the cAMP-binding domain of EPAC1 (DeltacAMP-EPAC1) appeared to disrupt targeting of EPAC1 in cells resulting in a more dispersed staining pattern. Using two-hybrid assay, we tested the ability of LC2 to interact with DeltacAMP-EPAC1 and DeltaDEP-EPAC1, which lacks a DEP domain (dishevelled, Egl-10 and pleckstrin homology domain). We found that deletion of the cAMP-binding domain inhibited interaction between EPAC1 and LC2 in a two-hybrid assay, but removal of the DEP domain had little effect. LC2 was found to interact with a glutathione-S-transferase-fusion protein of the cAMP-binding domain of EPAC1 in a pull-down assay, but not the DEP, REM (Ras exchange motif) or CAT (catalytic) domains. Together with our two-hybrid results, this suggests that the cAMP-binding domain of EPAC1 mediates interaction with LC2.
Figures

Similar articles
-
MAP1A light chain 2 interacts with exchange protein activated by cyclic AMP 1 (EPAC1) to enhance Rap1 GTPase activity and cell adhesion.J Biol Chem. 2005 Mar 4;280(9):8109-16. doi: 10.1074/jbc.M413697200. Epub 2004 Dec 9. J Biol Chem. 2005. PMID: 15591041
-
Dynamic interaction of cAMP with the Rap guanine-nucleotide exchange factor Epac1.J Mol Biol. 2001 Mar 9;306(5):1167-77. doi: 10.1006/jmbi.2001.4444. J Mol Biol. 2001. PMID: 11237625
-
Microtubule-associated protein 1B-light chain 1 enhances activation of Rap1 by exchange protein activated by cyclic AMP but not intracellular targeting.Mol Pharmacol. 2006 Jan;69(1):374-84. doi: 10.1124/mol.105.016337. Epub 2005 Oct 21. Mol Pharmacol. 2006. PMID: 16244178
-
Epac proteins: multi-purpose cAMP targets.Trends Biochem Sci. 2006 Dec;31(12):680-6. doi: 10.1016/j.tibs.2006.10.002. Epub 2006 Nov 2. Trends Biochem Sci. 2006. PMID: 17084085 Review.
-
Microtubule-associated proteins (MAPs) regulate cAMP signalling through exchange protein directly activated by cAMP (EPAC).Biochem Soc Trans. 2005 Dec;33(Pt 6):1327-9. doi: 10.1042/BST0331327. Biochem Soc Trans. 2005. PMID: 16246110 Review.
Cited by
-
The interaction of Epac1 and Ran promotes Rap1 activation at the nuclear envelope.Mol Cell Biol. 2010 Aug;30(16):3956-69. doi: 10.1128/MCB.00242-10. Epub 2010 Jun 14. Mol Cell Biol. 2010. PMID: 20547757 Free PMC article.
-
Epac: effectors and biological functions.Naunyn Schmiedebergs Arch Pharmacol. 2008 Jun;377(4-6):345-57. doi: 10.1007/s00210-007-0246-7. Epub 2008 Jan 5. Naunyn Schmiedebergs Arch Pharmacol. 2008. PMID: 18176800 Review.
-
Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing.Proc Natl Acad Sci U S A. 2014 Nov 4;111(44):E4726-35. doi: 10.1073/pnas.1404656111. Epub 2014 Oct 22. Proc Natl Acad Sci U S A. 2014. PMID: 25339441 Free PMC article.
-
Prostaglandin E2 inhibits specific lung fibroblast functions via selective actions of PKA and Epac-1.Am J Respir Cell Mol Biol. 2008 Oct;39(4):482-9. doi: 10.1165/rcmb.2008-0080OC. Epub 2008 Apr 17. Am J Respir Cell Mol Biol. 2008. PMID: 18421013 Free PMC article.
-
Genome-Wide Mapping Defines a Role for C/EBPβ and c-Jun in Non-Canonical Cyclic AMP Signalling.Cells. 2019 Oct 14;8(10):1253. doi: 10.3390/cells8101253. Cells. 2019. PMID: 31615122 Free PMC article.
References
-
- Kopperud R., Krakstad C., Selheim F., Doskeland S. O. cAMP effector mechanisms. Novel twists for an ‘old’ signaling system. FEBS Lett. 2003;546:121–126. - PubMed
-
- Houslay M. D. Adaptation in cyclic AMP signalling processes: a central role for cyclic AMP phosphodiesterases. Semin. Cell Dev. Biol. 1998;9:161–167. - PubMed
-
- Engh R. A., Bossemeyer D. The protein kinase activity modulation sites: mechanisms for cellular regulation – targets for therapeutic intervention. Adv. Enzyme Regul. 2001;41:121–149. - PubMed
-
- Medina D. L., Santisteban P. Thyrotropin-dependent proliferation of in vitro rat thyroid cell systems. Eur. J. Endocrinol. 2000;143:161–178. - PubMed
-
- Stork P. J., Schmitt J. M. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12:258–266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

