Microarray-based analysis of the Staphylococcus aureus sigmaB regulon
- PMID: 15205410
- PMCID: PMC421609
- DOI: 10.1128/JB.186.13.4085-4099.2004
Microarray-based analysis of the Staphylococcus aureus sigmaB regulon
Abstract
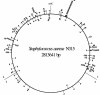
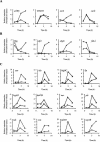
Microarray-based analysis of the transcriptional profiles of the genetically distinct Staphylococcus aureus strains COL, GP268, and Newman indicate that a total of 251 open reading frames (ORFs) are influenced by sigmaB activity. While sigmaB was found to positively control 198 genes by a factor of > or =2 in at least two of the three genetic lineages analyzed, 53 ORFs were repressed in the presence of sigmaB. Gene products that were found to be influenced by sigmaB are putatively involved in all manner of cellular processes, including cell envelope biosynthesis and turnover, intermediary metabolism, and signaling pathways. Most of the genes and/or operons identified as upregulated by sigmaB were preceded by a nucleotide sequence that resembled the sigmaB consensus promoter sequence of Bacillus subtilis. A conspicuous number of virulence-associated genes were identified as regulated by sigmaB activity, with many adhesins upregulated and prominently represented in this group, while transcription of various exoproteins and toxins were repressed. The data presented here suggest that the sigmaB of S. aureus controls a large regulon and is an important modulator of virulence gene expression that is likely to act conversely to RNAIII, the effector molecule of the agr locus. We propose that this alternative transcription factor may be of importance for the invading pathogen to fine-tune its virulence factor production in response to changing host environments.
Figures
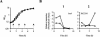

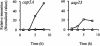
References
-
- Borukhov, S., and K. Severinov. 2002. Role of the RNA polymerase sigma subunit in transcription initiation. Res. Microbiol. 153:557-562. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

