The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica
- PMID: 15205413
- PMCID: PMC421605
- DOI: 10.1128/JB.186.13.4124-4133.2004
The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica
Abstract
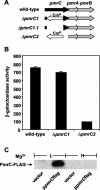
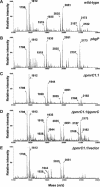
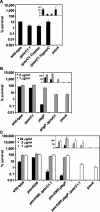
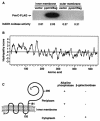
The PmrA/PmrB regulatory system of Salmonella enterica controls the modification of lipid A with aminoarabinose and phosphoethanolamine. The aminoarabinose modification is required for resistance to the antibiotic polymyxin B, as mutations of the PmrA-activated pbg operon or ugd gene result in strains that lack aminoarabinose in their lipid A molecules and are more susceptible to polymyxin B. Additional PmrA-regulated genes appear to participate in polymyxin B resistance, as pbgP and ugd mutants are not as sensitive to polymyxin B as a pmrA mutant. Moreover, the role that the phosphoethanolamine modification of lipid A plays in the resistance to polymyxin B has remained unknown. Here we address both of these questions by establishing that the PmrA-activated pmrC gene encodes an inner membrane protein that is required for the incorporation of phosphoethanolamine into lipid A and for polymyxin B resistance. The PmrC protein consists of an N-terminal region with five transmembrane domains followed by a large periplasmic region harboring the putative enzymatic domain. A pbgP pmrC double mutant resembled a pmrA mutant both in its lipid A profile and in its susceptibility to polymyxin B, indicating that the PmrA-dependent modification of lipid A with aminoarabinose and phosphoethanolamine is responsible for PmrA-regulated polymyxin B resistance.
Figures

Similar articles
-
Release of the lipopolysaccharide deacylase PagL from latency compensates for a lack of lipopolysaccharide aminoarabinose modification-dependent resistance to the antimicrobial peptide polymyxin B in Salmonella enterica.J Bacteriol. 2007 Jul;189(13):4911-9. doi: 10.1128/JB.00451-07. Epub 2007 May 4. J Bacteriol. 2007. PMID: 17483225 Free PMC article.
-
pmrA(Con) confers pmrHFIJKL-dependent EGTA and polymyxin resistance on msbB Salmonella by decorating lipid A with phosphoethanolamine.J Bacteriol. 2007 Jul;189(14):5161-9. doi: 10.1128/JB.01969-06. Epub 2007 Apr 20. J Bacteriol. 2007. PMID: 17449614 Free PMC article.
-
Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar typhimurium lipopolysaccharide core.J Bacteriol. 2005 May;187(10):3391-9. doi: 10.1128/JB.187.10.3391-3399.2005. J Bacteriol. 2005. PMID: 15866924 Free PMC article.
-
Lipid A Phosphoethanolamine Transferase: Regulation, Structure and Immune Response.J Mol Biol. 2020 Aug 21;432(18):5184-5196. doi: 10.1016/j.jmb.2020.04.022. Epub 2020 Apr 27. J Mol Biol. 2020. PMID: 32353363 Review.
-
Genetic and Biochemical Mechanisms for Bacterial Lipid A Modifiers Associated with Polymyxin Resistance.Trends Biochem Sci. 2019 Nov;44(11):973-988. doi: 10.1016/j.tibs.2019.06.002. Epub 2019 Jul 3. Trends Biochem Sci. 2019. PMID: 31279652 Review.
Cited by
-
Stereochemical Trajectories of a Two-Component Regulatory System PmrA/B in a Colistin-Resistant Acinetobacter baumannii Clinical Isolate.Iran Biomed J. 2021 May 1;25(3):193-201. doi: 10.29252/ibj.25.3.157. Iran Biomed J. 2021. PMID: 33653023 Free PMC article.
-
Combined transcriptomic and metabolomic analysis of Salmonella in the presence or absence of PhoP-PhoQ system under low Mg2+ conditions.Metabolomics. 2022 Nov 15;18(11):93. doi: 10.1007/s11306-022-01946-z. Metabolomics. 2022. PMID: 36378357
-
Colistin heteroresistance in Enterobacter cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy-l-arabinose addition to lipid A.Mol Microbiol. 2019 Jun;111(6):1604-1616. doi: 10.1111/mmi.14240. Epub 2019 Apr 10. Mol Microbiol. 2019. PMID: 30873646 Free PMC article.
-
Virulence Gene Profile, Antimicrobial Resistance and Multilocus Sequence Typing of Salmonella enterica Subsp. enterica Serovar Enteritidis from Chickens and Chicken Products.Animals (Basel). 2022 Jan 1;12(1):97. doi: 10.3390/ani12010097. Animals (Basel). 2022. PMID: 35011203 Free PMC article.
-
Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system.Antimicrob Agents Chemother. 2009 Sep;53(9):3628-34. doi: 10.1128/AAC.00284-09. Epub 2009 Jun 15. Antimicrob Agents Chemother. 2009. PMID: 19528270 Free PMC article.
References
-
- Bensadoun, A., and D. Weinstein. 1976. Assay of proteins in the presence of interfering materials. Anal. Biochem. 70:241-250. - PubMed
-
- Chamnongpol, S., W. Dodson, M. J. Cromie, Z. L. Harris, and E. A. Groisman. 2002. Fe(III)-mediated cellular toxicity. Mol. Microbiol. 45:711-719. - PubMed
-
- Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

