Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EsigmaA- and EsigmaE-responsive promoters by phosphorylated PhoP
- PMID: 15205429
- PMCID: PMC421599
- DOI: 10.1128/JB.186.13.4262-4275.2004
Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EsigmaA- and EsigmaE-responsive promoters by phosphorylated PhoP
Abstract
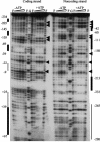
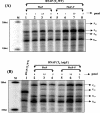
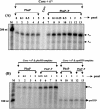
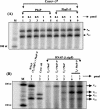
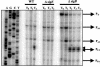
The phoPR operon encodes a response regulator, PhoP, and a histidine kinase, PhoR, which activate or repress genes of the Bacillus subtilis Pho regulon in response to an extracellular phosphate deficiency. Induction of phoPR upon phosphate starvation required activity of both PhoP and PhoR, suggesting autoregulation of the operon, a suggestion that is supported here by PhoP footprinting on the phoPR promoter. Primer extension analyses, using RNA from JH642 or isogenic sigE or sigB mutants isolated at different stages of growth and/or under different growth conditions, suggested that expression of the phoPR operon represents the sum of five promoters, each responding to a specific growth phase and environmental controls. The temporal expression of the phoPR promoters was investigated using in vitro transcription assays with RNA polymerase holoenzyme isolated at different stages of Pho induction, from JH642 or isogenic sigE or sigB mutants. In vitro transcription studies using reconstituted EsigmaA, EsigmaB, and EsigmaE holoenzymes identified PA4 and PA3 as EsigmaA promoters and PE2 as an EsigmaE promoter. Phosphorylated PhoP (PhoP approximately P) enhanced transcription from each of these promoters. EsigmaB was sufficient for in vitro transcription of the PB1 promoter. P5 was active only in a sigB mutant strain. These studies are the first to report a role for PhoP approximately P in activation of promoters that also have activity in the absence of Pho regulon induction and an activation role for PhoP approximately P at an EsigmaE promoter. Information concerning PB1 and P5 creates a basis for further exploration of the regulatory coordination or overlap of the PhoPR and SigB regulons during phosphate starvation.
Figures



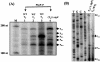
Similar articles
-
Transcriptional regulation of the phoPR operon in Bacillus subtilis.J Bacteriol. 2004 Feb;186(4):1182-90. doi: 10.1128/JB.186.4.1182-1190.2004. J Bacteriol. 2004. PMID: 14762014 Free PMC article.
-
Bacillus subtilis phosphorylated PhoP: direct activation of the E(sigma)A- and repression of the E(sigma)E-responsive phoB-PS+V promoters during pho response.J Bacteriol. 2005 Aug;187(15):5166-78. doi: 10.1128/JB.187.15.5166-5178.2005. J Bacteriol. 2005. PMID: 16030210 Free PMC article.
-
PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro.Mol Microbiol. 1998 Jun;28(6):1187-97. doi: 10.1046/j.1365-2958.1998.00882.x. Mol Microbiol. 1998. PMID: 9680208
-
The signal-transduction network for Pho regulation in Bacillus subtilis.Mol Microbiol. 1996 Mar;19(5):933-9. doi: 10.1046/j.1365-2958.1996.421953.x. Mol Microbiol. 1996. PMID: 8830274 Review.
-
Transcriptomic studies of phosphate control of primary and secondary metabolism in Streptomyces coelicolor.Appl Microbiol Biotechnol. 2012 Jul;95(1):61-75. doi: 10.1007/s00253-012-4129-6. Epub 2012 May 24. Appl Microbiol Biotechnol. 2012. PMID: 22622839 Review.
Cited by
-
Coupling between feedback loops in autoregulatory networks affects bistability range, open-loop gain and switching times.Phys Biol. 2012 Oct;9(5):055003. doi: 10.1088/1478-3975/9/5/055003. Epub 2012 Sep 25. Phys Biol. 2012. PMID: 23011599 Free PMC article.
-
Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis.J Bacteriol. 2005 Dec;187(23):8063-80. doi: 10.1128/JB.187.23.8063-8080.2005. J Bacteriol. 2005. PMID: 16291680 Free PMC article.
-
Dual role of the PhoP approximately P response regulator: Bacillus amyloliquefaciens FZB45 phytase gene transcription is directed by positive and negative interactions with the phyC promoter.J Bacteriol. 2006 Oct;188(19):6953-65. doi: 10.1128/JB.00681-06. J Bacteriol. 2006. PMID: 16980498 Free PMC article.
-
The two-component system PhoPR of Clostridium acetobutylicum is involved in phosphate-dependent gene regulation.J Bacteriol. 2008 Oct;190(20):6559-67. doi: 10.1128/JB.00574-08. Epub 2008 Aug 8. J Bacteriol. 2008. PMID: 18689481 Free PMC article.
-
Genome-wide analysis of phosphorylated PhoP binding to chromosomal DNA reveals several novel features of the PhoPR-mediated phosphate limitation response in Bacillus subtilis.J Bacteriol. 2015 Apr;197(8):1492-506. doi: 10.1128/JB.02570-14. Epub 2015 Feb 9. J Bacteriol. 2015. PMID: 25666134 Free PMC article.
References
-
- Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis and turnover, p. 381-410. In J. A. Hoch, R. Losick, and A. L. Sonenshein (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular biology. American Society for Microbiology, Washington, D.C.
-
- Blencke, H. M., G. Homuth, H. Ludwig, U. Mader, M. Hecker, and J. Stulke. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5:133-149. - PubMed
-
- Chesnut, R. S., C. Bookstein, and F. M. Hulett. 1991. Separate promoters direct expression of phoAIII, a member of the Bacillus subtilis alkaline phosphatase multigene family, during phosphate starvation and sporulation. Mol. Microbiol. 5:2181-2190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

