An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway
- PMID: 15205481
- PMCID: PMC470766
- DOI: 10.1073/pnas.0403166101
An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway
Abstract
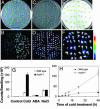
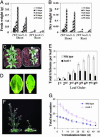
To investigate essential components mediating stress signaling in plants, we initiated a large-scale stress response screen using Arabidopsis plants carrying the firefly luciferase reporter gene under the control of the stress-responsive RD29A promoter. Here we report the identification and characterization of a mutant, hos9-1 (for high expression of osmotically responsive genes), in which the reporter construct was hyperactivated by low temperature, but not by abscisic acid or salinity stress. The mutants grow more slowly, and flower later, than do wild-type plants and are more sensitive to freezing, both before and after acclimation, than the wild-type plants. The HOS9 gene encodes a putative homeodomain transcription factor that is localized to the nucleus. HOS9 is constitutively expressed and not further induced by cold stress. Cold treatment increased the level of transcripts of the endogenous RD29A, and some other stress-responsive genes, to a higher level in hos9-1 than in wild-type plants. However, the C repeat/dehydration responsive element-binding factor (CBF) transcription factor genes that mediate a part of cold acclimation in Arabidopsis did not have their response to cold altered by the hos9-1 mutation. Correspondingly, microarray analysis showed that none of the genes affected by the hos9-1 mutation are controlled by the CBF family. Together, these results suggest that HOS9 is important for plant growth and development, and for a part of freezing tolerance, by affecting the activity of genes independent of the CBF pathway.
Figures




Similar articles
-
Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis.Plant Cell. 2003 Dec;15(12):2940-51. doi: 10.1105/tpc.015248. Epub 2003 Nov 20. Plant Cell. 2003. PMID: 14630965 Free PMC article.
-
Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis.Plant Physiol. 2002 Oct;130(2):639-48. doi: 10.1104/pp.006478. Plant Physiol. 2002. PMID: 12376631 Free PMC article.
-
Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis.Plant Physiol. 2016 Aug;171(4):2744-59. doi: 10.1104/pp.16.00533. Epub 2016 Jun 1. Plant Physiol. 2016. PMID: 27252305 Free PMC article.
-
CBF-dependent signaling pathway: a key responder to low temperature stress in plants.Crit Rev Biotechnol. 2011 Jun;31(2):186-92. doi: 10.3109/07388551.2010.505910. Epub 2010 Oct 4. Crit Rev Biotechnol. 2011. PMID: 20919819 Review.
-
Molecular genetic analysis of cold-regulated gene transcription.Philos Trans R Soc Lond B Biol Sci. 2002 Jul 29;357(1423):877-86. doi: 10.1098/rstb.2002.1076. Philos Trans R Soc Lond B Biol Sci. 2002. PMID: 12171651 Free PMC article. Review.
Cited by
-
The sensitive to freezing3 mutation of Arabidopsis thaliana is a cold-sensitive allele of homomeric acetyl-CoA carboxylase that results in cold-induced cuticle deficiencies.J Exp Bot. 2012 Sep;63(14):5289-99. doi: 10.1093/jxb/ers191. Epub 2012 Jul 12. J Exp Bot. 2012. PMID: 22791831 Free PMC article.
-
Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis.Plant Cell Rep. 2005 Dec;24(11):683-90. doi: 10.1007/s00299-005-0061-x. Epub 2005 Oct 18. Plant Cell Rep. 2005. PMID: 16231185
-
Cold transiently activates calcium-permeable channels in Arabidopsis mesophyll cells.Plant Physiol. 2007 Jan;143(1):487-94. doi: 10.1104/pp.106.090928. Epub 2006 Nov 17. Plant Physiol. 2007. PMID: 17114272 Free PMC article.
-
SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis.Plant Cell. 2007 Apr;19(4):1403-14. doi: 10.1105/tpc.106.048397. Epub 2007 Apr 6. Plant Cell. 2007. PMID: 17416732 Free PMC article.
-
Characterization and expression profiles of WUSCHEL-related homeobox (WOX) gene family in cultivated alfalfa (Medicago sativa L.).BMC Plant Biol. 2023 Oct 6;23(1):471. doi: 10.1186/s12870-023-04476-5. BMC Plant Biol. 2023. PMID: 37803258 Free PMC article.
References
-
- Guy, C. L. (1990) Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 187-223.
-
- Hughes, M. A. & Dunn, M. A. (1996) J. Exp. Bot. 47, 291-305.
-
- Thomashow, M. F. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571-599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

