Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes
- PMID: 15208399
- PMCID: PMC514152
- DOI: 10.1105/tpc.021345
Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes
Abstract
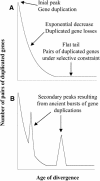
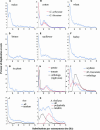
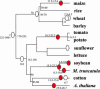
It is often anticipated that many of today's diploid plant species are in fact paleopolyploids. Given that an ancient large-scale duplication will result in an excess of relatively old duplicated genes with similar ages, we analyzed the timing of duplication of pairs of paralogous genes in 14 model plant species. Using EST contigs (unigenes), we identified pairs of paralogous genes in each species and used the level of synonymous nucleotide substitution to estimate the relative ages of gene duplication. For nine of the investigated species (wheat [Triticum aestivum], maize [Zea mays], tetraploid cotton [Gossypium hirsutum], diploid cotton [G. arboretum], tomato [Lycopersicon esculentum], potato [Solanum tuberosum], soybean [Glycine max], barrel medic [Medicago truncatula], and Arabidopsis thaliana), the age distributions of duplicated genes contain peaks corresponding to short evolutionary periods during which large numbers of duplicated genes were accumulated. Large-scale duplications (polyploidy or aneuploidy) are strongly suspected to be the cause of these temporal peaks of gene duplication. However, the unusual age profile of tandem gene duplications in Arabidopsis indicates that other scenarios, such as variation in the rate at which duplicated genes are deleted, must also be considered.
Figures
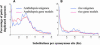
Comment in
-
Two genomes are better than one: widespread paleopolyploidy in plants and evolutionary effects.Plant Cell. 2004 Jul;16(7):1647-9. doi: 10.1105/tpc.160710. Plant Cell. 2004. PMID: 15272471 Free PMC article. Review. No abstract available.
References
-
- Adams, M.D., et al. (1991). Complementary DNA sequencing: Expressed sequence tags and human genome project. Science 252, 1651–1656. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Aoki, N., Whitfeld, P., Hoeren, F., Scofield, G., Newell, K., Patrick, J., Offler, C., Clarke, B., Rahman, S., and Furbank, R.T. (2002). Three sucrose transporter genes are expressed in the developing grain of hexaploid wheat. Plant Mol. Biol. 50, 453–462. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

