The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes
- PMID: 15208421
- PMCID: PMC514111
- DOI: 10.1104/pp.104.040030
The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes
Abstract
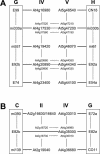
The annotated Arabidopsis genome sequence was exploited as a tool for carrying out comparative analyses of the Arabidopsis and Capsella rubella genomes. Comparison of a set of random, short C. rubella sequences with the corresponding sequences in Arabidopsis revealed that aligned protein-coding exon sequences differ from aligned intron or intergenic sequences in respect to the degree of sequence identity and the frequency of small insertions/deletions. Molecular-mapped markers and expressed sequence tags derived from Arabidopsis were used for genetic mapping in a population derived from an interspecific cross between Capsella grandiflora and C. rubella. The resulting eight Capsella linkage groups were compared to the sequence maps of the five Arabidopsis chromosomes. Fourteen colinear segments spanning approximately 85% of the Arabidopsis chromosome sequence maps and 92% of the Capsella genetic linkage map were detected. Several fusions and fissions of chromosomal segments as well as large inversions account for the observed arrangement of the 14 colinear blocks in the analyzed genomes. In addition, evidence for small-scale deviations from genome colinearity was found. Colinearity between the Arabidopsis and Capsella genomes is more pronounced than has been previously reported for comparisons between Arabidopsis and different Brassica species.
Figures


References
-
- Acarkan A, Rossberg M, Koch M, Schmidt R (2000) Comparative genome analysis reveals extensive conservation of genome organisation for Arabidopsis thaliana and Capsella rubella. Plant J 23: 55–62 - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Babula D, Kaczmarek M, Barakat A, Delseny M, Quiros CF, Sadowski J (2003) Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol Genet Genomics 268: 656–665 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

