Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot
- PMID: 15208422
- PMCID: PMC514113
- DOI: 10.1104/pp.104.039826
Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot
Abstract
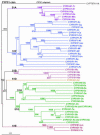
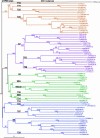
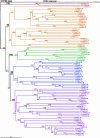
Data mining methods have been used to identify 356 Cyt P450 genes and 99 related pseudogenes in the rice (Oryza sativa) genome using sequence information available from both the indica and japonica strains. Because neither of these genomes is completely available, some genes have been identified in only one strain, and 28 genes remain incomplete. Comparison of these rice genes with the 246 P450 genes and 26 pseudogenes in the Arabidopsis genome has indicated that most of the known plant P450 families existed before the monocot-dicot divergence that occurred approximately 200 million years ago. Comparative analysis of P450s in the Pinus expressed sequence tag collections has identified P450 families that predated the separation of gymnosperms and flowering plants. Complete mapping of all available plant P450s onto the Deep Green consensus plant phylogeny highlights certain lineage-specific families maintained (CYP80 in Ranunculales) and lineage-specific families lost (CYP92 in Arabidopsis) in the course of evolution.
Figures



Similar articles
-
Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining.BMC Genomics. 2008 Feb 21;9:86. doi: 10.1186/1471-2164-9-86. BMC Genomics. 2008. PMID: 18291034 Free PMC article.
-
Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice.Plant Cell. 2007 Aug;19(8):2329-48. doi: 10.1105/tpc.107.051300. Epub 2007 Aug 24. Plant Cell. 2007. PMID: 17720868 Free PMC article.
-
Evolutionary history and functional divergence of the cytochrome P450 gene superfamily between Arabidopsis thaliana and Brassica species uncover effects of whole genome and tandem duplications.BMC Genomics. 2017 Sep 18;18(1):733. doi: 10.1186/s12864-017-4094-7. BMC Genomics. 2017. PMID: 28923019 Free PMC article.
-
Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis.Plant Cell Physiol. 2004 Sep;45(9):1111-21. doi: 10.1093/pcp/pch151. Plant Cell Physiol. 2004. PMID: 15509833 Review.
-
Exploring the genomes: from Arabidopsis to crops.J Plant Physiol. 2011 Jan 1;168(1):3-8. doi: 10.1016/j.jplph.2010.07.008. J Plant Physiol. 2011. PMID: 20817312 Review.
Cited by
-
Oryza sativa cytochrome P450 family member OsCYP96B4 reduces plant height in a transcript dosage dependent manner.PLoS One. 2011;6(11):e28069. doi: 10.1371/journal.pone.0028069. Epub 2011 Nov 28. PLoS One. 2011. PMID: 22140509 Free PMC article.
-
A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.).Plant Mol Biol. 2015 May;88(1-2):85-99. doi: 10.1007/s11103-015-0310-5. Epub 2015 Mar 24. Plant Mol Biol. 2015. PMID: 25800365
-
Overexpression of the PanaxginsengCYP703 Alters Cutin Composition of Reproductive Tissues in Arabidopsis.Plants (Basel). 2022 Jan 30;11(3):383. doi: 10.3390/plants11030383. Plants (Basel). 2022. PMID: 35161364 Free PMC article.
-
A new lead chemical for strigolactone biosynthesis inhibitors.Plant Cell Physiol. 2010 Jul;51(7):1143-50. doi: 10.1093/pcp/pcq077. Epub 2010 Jun 3. Plant Cell Physiol. 2010. PMID: 20522488 Free PMC article.
-
Computational evidence for hundreds of non-conserved plant microRNAs.BMC Genomics. 2005 Sep 13;6:119. doi: 10.1186/1471-2164-6-119. BMC Genomics. 2005. PMID: 16159385 Free PMC article.
References
-
- Agrawal GK, Tamogami S, Han O, Iwahashi H, Rakwal R (2004) Rice octadecanoid pathway. Biochem Biophys Res Commun 317: 1–15 - PubMed
-
- Akashi T, Fukuchi-Mizutani M, Aoki T, Ueyama Y, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Ayabe S (1999) Molecular cloning and biochemical characterization of a novel cytochrome P450, flavone synthase II, that catalyzes direct conversion of flavanones to flavones. Plant Cell Physiol 40: 1182–1186 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

