Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes
- PMID: 15208423
- PMCID: PMC514114
- DOI: 10.1104/pp.104.042176
Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes
Abstract
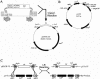
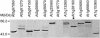
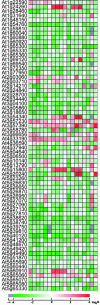
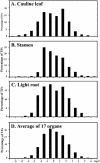
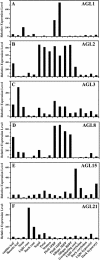
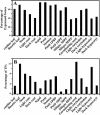
Here, we report our effort in generating an ORFeome collection for the Arabidopsis transcription factor (TF) genes. In total, ORFeome clones representing 1,282 Arabidopsis TF genes have been obtained in the Gateway high throughput cloning pENTR vector, including 411 genes whose annotation lack cDNA support. All the ORFeome inserts have also been mobilized into a yeast expression destination vector, with an estimated 85% rate of expressing the respective proteins. Sequence analysis of these clones revealed that 34 of them did not match with either the reported cDNAs or current predicted open-reading-frame sequences. Among those, novel alternative splicing of TF gene transcripts is responsible for the observed differences in at least five genes. However, those alternative splicing events do not appear to be differentially regulated among distinct Arabidopsis tissues examined. Lastly, expression of those TF genes in 17 distinct Arabidopsis organ types and the cultured cells was profiled using a 70-mer oligo microarray.
Figures

References
-
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Burgeff C, Liljegren SJ, Tapia-López R, Yanofsky MF, Alvarez-Buylla ER (2002) MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214: 365–372 - PubMed
-
- Flanagan CA, Hu Y, Ma H (1996) Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J 10: 343–353 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

