Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses
- PMID: 15208424
- PMCID: PMC514115
- DOI: 10.1104/pp.103.035584
Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses
Abstract
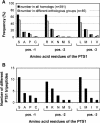
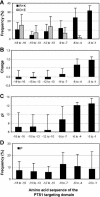
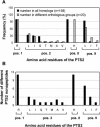
To specify the C-terminal peroxisome targeting signal type 1 (PTS1) and the N-terminal PTS2 for higher plants, a maximum number of plant cDNAs and expressed sequence tags that are homologous to PTS1- and PTS2-targeted plant proteins was retrieved from the public databases and the primary structure of their targeting domains was analyzed for conserved properties. According to their high overall frequency in the homologs and their widespread occurence in different orthologous groups, nine major PTS1 tripeptides ([SA][RK][LM]> without AKM> plus SRI> and PRL>) and two major PTS2 nonapeptides (R[LI]x5HL) were defined that are considered good indicators for peroxisomal localization if present in unknown proteins. A lower but significant number of homologs contained 1 of 11 minor PTS1 tripeptides or of 9 minor PTS2 nonapeptides, many of which have not been identified before in plant peroxisomal proteins. The region adjacent to the PTS peptides was characterized by specific conserved properties as well, such as a pronounced incidence of basic and Pro residues and a high positive net charge, which probably play an auxiliary role in peroxisomal targeting. By contrast, several peptides with assumed peroxisomal targeting properties were not found in any of the 550 homologs and hence play--if at all--only a minor role in peroxisomal targeting. Based on the definition of these major and minor PTS and on the recognition of additional conserved properties, the accuracy of predicting peroxisomal proteins can be raised and plant genomes can be screened for novel proteins of peroxisomes more successfully.
Figures

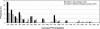




Similar articles
-
The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5.Mol Biol Cell. 2005 Feb;16(2):573-83. doi: 10.1091/mbc.e04-05-0422. Epub 2004 Nov 17. Mol Biol Cell. 2005. PMID: 15548601 Free PMC article.
-
Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import.Mol Cell Biol. 2002 Mar;22(6):1639-55. doi: 10.1128/MCB.22.6.1639-1655.2002. Mol Cell Biol. 2002. PMID: 11865044 Free PMC article.
-
PTS1 peroxisomal import pathway plays shared and distinct roles to PTS2 pathway in development and pathogenicity of Magnaporthe oryzae.PLoS One. 2013;8(2):e55554. doi: 10.1371/journal.pone.0055554. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405169 Free PMC article.
-
Characterization, prediction and evolution of plant peroxisomal targeting signals type 1 (PTS1s).Biochim Biophys Acta. 2016 May;1863(5):790-803. doi: 10.1016/j.bbamcr.2016.01.001. Epub 2016 Jan 6. Biochim Biophys Acta. 2016. PMID: 26772785 Review.
-
Entering a new era of research on plant peroxisomes.Curr Opin Plant Biol. 2003 Dec;6(6):577-82. doi: 10.1016/j.pbi.2003.09.012. Curr Opin Plant Biol. 2003. PMID: 14611956 Review.
Cited by
-
Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers.Plant Cell. 2012 May;24(5):2015-30. doi: 10.1105/tpc.112.097519. Epub 2012 May 30. Plant Cell. 2012. PMID: 22649270 Free PMC article.
-
AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes.Plant Physiol. 2004 Sep;136(1):2587-608. doi: 10.1104/pp.104.043695. Epub 2004 Aug 27. Plant Physiol. 2004. PMID: 15333753 Free PMC article.
-
Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery.Plant Physiol. 2007 Jun;144(2):1132-43. doi: 10.1104/pp.107.098897. Epub 2007 Apr 20. Plant Physiol. 2007. PMID: 17449647 Free PMC article.
-
Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions.Plant Physiol. 2005 Aug;138(4):2111-23. doi: 10.1104/pp.105.066225. Epub 2005 Jul 29. Plant Physiol. 2005. PMID: 16055677 Free PMC article.
-
Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice.Plant Mol Biol. 2008 Nov;68(4-5):439-49. doi: 10.1007/s11103-008-9381-x. Epub 2008 Aug 13. Plant Mol Biol. 2008. PMID: 18704694
References
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Dammai V, Subramani S (2001) The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105: 187–196 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

