Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins
- PMID: 15208425
- PMCID: PMC514142
- DOI: 10.1104/pp.104.040436
Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins
Abstract
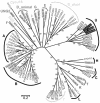
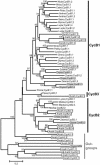
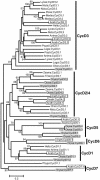
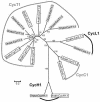
Cyclins are primary regulators of the activity of cyclin-dependent kinases, which are known to play critical roles in controlling eukaryotic cell cycle progression. While there has been extensive research on cell cycle mechanisms and cyclin function in animals and yeasts, only a small number of plant cyclins have been characterized functionally. In this paper, we describe an exhaustive search for cyclin genes in the Arabidopsis genome and among available sequences from other vascular plants. Based on phylogenetic analysis, we define 10 classes of plant cyclins, four of which are plant-specific, and a fifth is shared between plants and protists but not animals. Microarray and reverse transcriptase-polymerase chain reaction analyses further provide expression profiles of cyclin genes in different tissues of wild-type Arabidopsis plants. Comparative phylogenetic studies of 174 plant cyclins were also performed. The phylogenetic results imply that the cyclin gene family in plants has experienced more gene duplication events than in animals. Expression patterns and phylogenetic analyses of Arabidopsis cyclin genes suggest potential gene redundancy among members belonging to the same group. We discuss possible divergence and conservation of some plant cyclins. Our study provides an opportunity to rapidly assess the position of plant cyclin genes in terms of evolution and classification, serving as a guide for further functional study of plant cyclins.
Figures



Similar articles
-
Genome-wide analysis of cyclin family in rice (Oryza Sativa L.).Mol Genet Genomics. 2006 Apr;275(4):374-86. doi: 10.1007/s00438-005-0093-5. Epub 2006 Jan 25. Mol Genet Genomics. 2006. PMID: 16435118
-
Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control.Plant J. 2005 Feb;41(4):546-66. doi: 10.1111/j.1365-313X.2004.02319.x. Plant J. 2005. PMID: 15686519
-
Genome-wide analysis of cyclins in maize (Zea mays).Genet Mol Res. 2010 Aug 3;9(3):1490-503. doi: 10.4238/vol9-3gmr861. Genet Mol Res. 2010. PMID: 20690081
-
Factors controlling cyclin B expression.Plant Mol Biol. 2000 Aug;43(5-6):677-90. doi: 10.1023/a:1006336005587. Plant Mol Biol. 2000. PMID: 11089869 Review.
-
Cell cycle controls: genome-wide analysis in Arabidopsis.Curr Opin Plant Biol. 2001 Dec;4(6):501-6. doi: 10.1016/s1369-5266(00)00207-7. Curr Opin Plant Biol. 2001. PMID: 11641065 Review.
Cited by
-
Genome-Wide Identification, Characterization, and Transcriptomic Analysis of the Cyclin Gene Family in Brassica rapa.Int J Mol Sci. 2022 Nov 13;23(22):14017. doi: 10.3390/ijms232214017. Int J Mol Sci. 2022. PMID: 36430495 Free PMC article.
-
Control of the meiotic cell division program in plants.Plant Reprod. 2013 Sep;26(3):143-58. doi: 10.1007/s00497-013-0223-x. Epub 2013 Jul 14. Plant Reprod. 2013. PMID: 23852379 Free PMC article. Review.
-
RNA-seq analysis of an apical meristem time series reveals a critical point in Arabidopsis thaliana flower initiation.BMC Genomics. 2015 Jun 18;16(1):466. doi: 10.1186/s12864-015-1688-9. BMC Genomics. 2015. PMID: 26084880 Free PMC article.
-
Genome-wide expression profiling and identification of gene activities during early flower development in Arabidopsis.Plant Mol Biol. 2005 Jun;58(3):401-19. doi: 10.1007/s11103-005-5434-6. Plant Mol Biol. 2005. PMID: 16021403
-
Repair of DNA double-strand breaks in plant meiosis: role of eukaryotic RecA recombinases and their modulators.Plant Reprod. 2023 Mar;36(1):17-41. doi: 10.1007/s00497-022-00443-6. Epub 2022 Jun 1. Plant Reprod. 2023. PMID: 35641832 Review.
References
-
- Abrahams S, Cavet G, Oakenfull EA, Carmichael JP, Shah ZH, Soni R, Murray JA (2001) A novel and highly divergent Arabidopsis cyclin isolated by complementation in budding yeast. Biochim Biophys Acta 1539: 1–6 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

