A horizontally acquired group II intron in the chloroplast psbA gene of a psychrophilic Chlamydomonas: in vitro self-splicing and genetic evidence for maturase activity
- PMID: 15208445
- PMCID: PMC1370600
- DOI: 10.1261/rna.7140604
A horizontally acquired group II intron in the chloroplast psbA gene of a psychrophilic Chlamydomonas: in vitro self-splicing and genetic evidence for maturase activity
Abstract
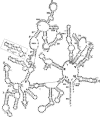
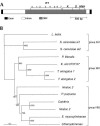
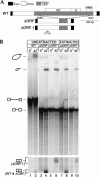
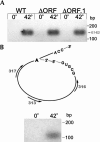
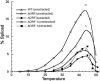
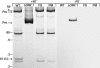
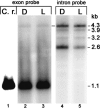
The majority of known group II introns are from chloroplast genomes, yet the first self-splicing group II intron from a chloroplast gene was reported only recently, from the psbA gene of the euglenoid, Euglena myxocylindracea. Herein, we describe a large (2.6-kb) group II intron from the psbA gene (psbA1) of a psychrophilic Chlamydomonas sp. from Antarctica that self-splices accurately in vitro. Remarkably, this intron, which also encodes an ORF with putative reverse transcriptase, maturase, and endonuclease domains, is in the same location, and is related to the E. myxocylindracea intron, as well as to group IIB2 introns from cyanobacteria. In vitro self-splicing of Chs.psbA1 occurred via a lariat, and required Mg(2+) (>12 mM) and NH(4)(+). Self-splicing was improved by deleting most of the ORF and by using pre-RNAs directly from transcription reactions, suggestive of a role for folding during transcription. Self-splicing of Chs.psbA1 pre-RNAs showed temperature optima of ~44 degrees C, but with a broad shoulder on the low side of the peak; splicing was nearly absent at 50 degrees C, indicative of thermolability. Splicing of wild-type Chs.psbA1 also occurred in Escherichia coli, but not when the ORF was disrupted by mutations, providing genetic evidence that it has maturase activity. This work provides the first description of a ribozyme from a psychrophilic organism. It also appears to provide a second instance of interkingdom horizontal transfer of this group IIB2 intron (or a close relative) from cyanobacteria to chloroplasts.
Figures

Similar articles
-
Recent horizontal intron transfer to a chloroplast genome.Nucleic Acids Res. 2004 Feb 3;32(2):803-10. doi: 10.1093/nar/gkh225. Print 2004. Nucleic Acids Res. 2004. PMID: 14762207 Free PMC article.
-
Self-splicing of the Chlamydomonas chloroplast psbA introns.Plant Cell. 1991 Oct;3(10):1095-107. doi: 10.1105/tpc.3.10.1095. Plant Cell. 1991. PMID: 1821761 Free PMC article.
-
Evidence for light/redox-regulated splicing of psbA pre-RNAs in Chlamydomonas chloroplasts.RNA. 1997 Jan;3(1):37-48. RNA. 1997. PMID: 8990397 Free PMC article.
-
Insights into the strategies used by related group II introns to adapt successfully for the colonisation of a bacterial genome.RNA Biol. 2014;11(8):1061-71. doi: 10.4161/rna.32092. Epub 2014 Oct 31. RNA Biol. 2014. PMID: 25482895 Free PMC article. Review.
-
Group II introns and expression of conjugative transfer functions in lactic acid bacteria.Antonie Van Leeuwenhoek. 1999 Jul-Nov;76(1-4):77-88. Antonie Van Leeuwenhoek. 1999. PMID: 10532373 Review.
Cited by
-
An evaluation of detection methods for large lariat RNAs.RNA. 2005 Mar;11(3):323-31. doi: 10.1261/rna.7124405. Epub 2005 Jan 20. RNA. 2005. PMID: 15661842 Free PMC article.
-
Multiple physical forms of excised group II intron RNAs in wheat mitochondria.Nucleic Acids Res. 2006 May 22;34(9):2782-90. doi: 10.1093/nar/gkl328. Print 2006. Nucleic Acids Res. 2006. PMID: 16717283 Free PMC article.
-
An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters.BMC Biol. 2006 Sep 6;4:31. doi: 10.1186/1741-7007-4-31. BMC Biol. 2006. PMID: 16956407 Free PMC article.
-
Categorizing 161 plant (streptophyte) mitochondrial group II introns into 29 families of related paralogues finds only limited links between intron mobility and intron-borne maturases.BMC Ecol Evol. 2023 Mar 13;23(1):5. doi: 10.1186/s12862-023-02108-y. BMC Ecol Evol. 2023. PMID: 36915058 Free PMC article.
-
Evolution of group II introns.Mob DNA. 2015 Apr 1;6:7. doi: 10.1186/s13100-015-0037-5. eCollection 2015. Mob DNA. 2015. PMID: 25960782 Free PMC article.
References
-
- Adamidi, C., Fedorova, O., and Pyle, A.M. 2003. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry 42: 3409–3418. - PubMed
-
- Bonen, L. and Vogel, J. 2001. The ins and outs of group II introns. Trends Genet. 17: 322–331. - PubMed
-
- Bunse, A.A., Nickelsen, J., and Kück, U. 2001. Intron-specific RNA binding proteins in the chloroplast of the green alga Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1519: 46–54. - PubMed
-
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BioMed Central Bioinform. 3: 2. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
