In silico prediction of peptides binding to multiple HLA-DR molecules accurately identifies immunodominant epitopes from gp43 of Paracoccidioides brasiliensis frequently recognized in primary peripheral blood mononuclear cell responses from sensitized individuals
- PMID: 15208742
- PMCID: PMC1430984
In silico prediction of peptides binding to multiple HLA-DR molecules accurately identifies immunodominant epitopes from gp43 of Paracoccidioides brasiliensis frequently recognized in primary peripheral blood mononuclear cell responses from sensitized individuals
Abstract
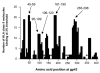
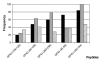
One of the major drawbacks limiting the use of synthetic peptide vaccines in genetically distinct populations is the fact that different epitopes are recognized by T cells from individuals displaying distinct major histocompatibility complex molecules. Immunization of mice with peptide (181-195) from the immunodominant 43 kDa glycoprotein of Paracoccidioides brasiliensis (gp43), the causative agent of Paracoccidioidomycosis (PCM), conferred protection against infectious challenge by the fungus. To identify immunodominant and potentially protective human T-cell epitopes in gp43, we used the TEPITOPE algorithm to select peptide sequences that would most likely bind multiple HLA-DR molecules and tested their recognition by T cells from sensitized individuals. The 5 most promiscuous peptides were selected from the gp43 sequence and the actual promiscuity of HLA binding was assessed by direct binding assays to 9 prevalent HLA-DR molecules. Synthetic peptides were tested in proliferation assays with peripheral blood mononuclear cells (PBMC) from PCM patients after chemotherapy and healthy controls. PBMC from 14 of 19 patients recognized at least one of the promiscuous peptides, whereas none of the healthy controls recognized the gp43 promiscuous peptides. Peptide gp43(180-194) was recognized by 53% of patients, whereas the other promiscuous gp43 peptides were recognized by 32% to 47% of patients. The frequency of peptide binding and peptide recognition correlated with the promiscuity of HLA-DR binding, as determined by TEPITOPE analysis. In silico prediction of promiscuous epitopes led to the identification of naturally immunodominant epitopes recognized by PBMC from a significant proportion of a genetically heterogeneous patient population exposed to P. brasiliensis. The combination of several such epitopes may increase the frequency of positive responses and allow the immunization of genetically distinct populations.
Figures


Similar articles
-
T-cell recognition of Paracoccidioides brasiliensis gp43-derived peptides in patients with paracoccidioidomycosis and healthy individuals.Clin Vaccine Immunol. 2007 Apr;14(4):474-6. doi: 10.1128/CVI.00458-06. Epub 2007 Feb 28. Clin Vaccine Immunol. 2007. PMID: 17329443 Free PMC article.
-
Identification of novel consensus CD4 T-cell epitopes from clade B HIV-1 whole genome that are frequently recognized by HIV-1 infected patients.AIDS. 2006 Nov 28;20(18):2263-73. doi: 10.1097/01.aids.0000253353.48331.5f. AIDS. 2006. PMID: 17117012
-
Synthesis and immunological activity of a branched peptide carrying the T-cell epitope of gp43, the major exocellular antigen of Paracoccidioides brasiliensis.Scand J Immunol. 2004 Jan;59(1):58-65. doi: 10.1111/j.0300-9475.2004.01359.x. Scand J Immunol. 2004. PMID: 14723622
-
Attempts at a peptide vaccine against paracoccidioidomycosis, adjuvant to chemotherapy.Mycopathologia. 2008 Apr-May;165(4-5):341-52. doi: 10.1007/s11046-007-9056-1. Mycopathologia. 2008. PMID: 18777638 Review.
-
Biochemistry and molecular biology of the main diagnostic antigen of Paracoccidioides brasiliensis.Arch Med Res. 1995 Autumn;26(3):297-304. Arch Med Res. 1995. PMID: 8580684 Review.
Cited by
-
Multiple-Allele MHC Class II Epitope Engineering by a Molecular Dynamics-Based Evolution Protocol.Front Immunol. 2022 Apr 27;13:862851. doi: 10.3389/fimmu.2022.862851. eCollection 2022. Front Immunol. 2022. PMID: 35572587 Free PMC article.
-
CD4+ T cells from HIV-1-infected patients recognize wild-type and mutant human immunodeficiency virus-1 protease epitopes.Clin Exp Immunol. 2011 Apr;164(1):90-9. doi: 10.1111/j.1365-2249.2011.04319.x. Epub 2011 Feb 24. Clin Exp Immunol. 2011. PMID: 21352200 Free PMC article.
-
Connection between MHC class II binding and aggregation propensity: The antigenic peptide 10 of Paracoccidioides brasiliensis as a benchmark study.Comput Struct Biotechnol J. 2023 Feb 18;21:1746-1758. doi: 10.1016/j.csbj.2023.02.031. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 36890879 Free PMC article.
-
Characterization of conserved combined T and B cell epitopes in Leptospira interrogans major outer membrane proteins OmpL1 and LipL41.BMC Microbiol. 2011 Jan 26;11(1):21. doi: 10.1186/1471-2180-11-21. BMC Microbiol. 2011. PMID: 21269437 Free PMC article.
-
Epitope mapping of Acinetobacter baumannii outer membrane protein W (OmpW) and laboratory study of an OmpW-derivative peptide.Heliyon. 2023 Jul 25;9(8):e18614. doi: 10.1016/j.heliyon.2023.e18614. eCollection 2023 Aug. Heliyon. 2023. PMID: 37560650 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
