Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A
- PMID: 15210745
- PMCID: PMC2212815
- DOI: 10.1084/jem.20032202
Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A
Abstract
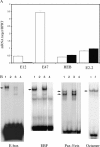
The basic helix-loop-helix transcription factors encoded by the E2A gene function at the apex of a transcriptional hierarchy involving E2A, early B cell factor (EBF), and Pax5, which is essential for B lymphopoiesis. In committed B lineage progenitors, E2A proteins have also been shown to regulate many lineage-associated genes. Herein, we demonstrate that the block in B lymphopoiesis imposed by the absence of E2A can be overcome by expression of EBF, but not Pax5, indicating that EBF is the essential target of E2A required for development of B lineage progenitors. Our data demonstrate that EBF, in synergy with low levels of alternative E2A-related proteins (E proteins), is sufficient to promote expression of most B lineage genes. Remarkably, however, we find that E2A proteins are required for interleukin 7-dependent proliferation due, in part, to a role for E2A in optimal expression of N-myc. Therefore, high levels of E protein activity are essential for the activation of EBF and N-myc, whereas lower levels of E protein activity, in synergy with other B lineage transcription factors, are sufficient for expression of most B lineage genes.
Figures




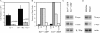

References
-
- Kee, B.L., and C.J. Paige. 1995. Murine B cell development: commitment and progression from multipotential progenitors to mature B lymphocytes. Int. Rev. Cytol. 157:129–179. - PubMed
-
- Kee, B.L., and C. Murre. 2001. Transcription factor regulation of B lineage commitment. Curr. Opin. Immunol. 13:180–185. - PubMed
-
- O'Riordan, M., and R. Grosschedl. 2000. Transcriptional regulation of early B-lymphocyte differentiation. Immunol. Rev. 175:94–103. - PubMed
-
- Bain, G., E.C. Robanus Maandag, D.J. Izon, D. Amsen, A.M. Kruisbeek, B.C. Weintraub, I. Krop, M.S. Schlissel, A.J. Feeney, M. van Roon, et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 79:885–892. - PubMed
-
- Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell. 79:875–884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

