The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae
- PMID: 15210978
- PMCID: PMC470741
- DOI: 10.1073/pnas.0305659101
The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae
Abstract
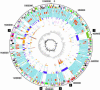
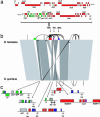
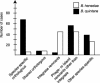
We present the complete genomes of two human pathogens, Bartonella quintana (1,581,384 bp) and Bartonella henselae (1,931,047 bp). The two pathogens maintain several similarities in being transmitted by insect vectors, using mammalian reservoirs, infecting similar cell types (endothelial cells and erythrocytes) and causing vasculoproliferative changes in immunocompromised hosts. A primary difference between the two pathogens is their reservoir ecology. Whereas B. quintana is a specialist, using only the human as a reservoir, B. henselae is more promiscuous and is frequently isolated from both cats and humans. Genome comparison elucidated a high degree of overall similarity with major differences being B. henselae specific genomic islands coding for filamentous hemagglutinin, and evidence of extensive genome reduction in B. quintana, reminiscent of that found in Rickettsia prowazekii. Both genomes are reduced versions of chromosome I from the highly related pathogen Brucella melitensis. Flanked by two rRNA operons is a segment with similarity to genes located on chromosome II of B. melitensis, suggesting that it was acquired by integration of megareplicon DNA in a common ancestor of the two Bartonella species. Comparisons of the vector-host ecology of these organisms suggest that the utilization of host-restricted vectors is associated with accelerated rates of genome degradation and may explain why human pathogens transmitted by specialist vectors are outnumbered by zoonotic agents, which use vectors of broad host ranges.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

