Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry
- PMID: 15210983
- PMCID: PMC470779
- DOI: 10.1073/pnas.0402700101
Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry
Abstract
Peptide sequence analysis using a combination of gas-phase ion/ion chemistry and tandem mass spectrometry (MS/MS) is demonstrated. Singly charged anthracene anions transfer an electron to multiply protonated peptides in a radio frequency quadrupole linear ion trap (QLT) and induce fragmentation of the peptide backbone along pathways that are analogous to those observed in electron capture dissociation. Modifications to the QLT that enable this ion/ion chemistry are presented, and automated acquisition of high-quality, single-scan electron transfer dissociation MS/MS spectra of phosphopeptides separated by nanoflow HPLC is described.
Figures
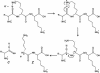
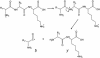
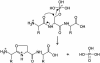




References
-
- Zubarev, R. A., Kelleher, N. L. & McLafferty, F. W. (1998) J. Am. Chem. Soc. 120, 3265-3266.
-
- Zubarev, R. A., Kruger, N. A., Fridriksson, E. K., Lewis, M. A., Horn, D. M., Carpenter, B. K. & McLafferty, F. W. (1999) J. Am. Chem. Soc. 121, 2857-2862.
-
- Zubarev, R. A. (2003) Mass Spectrom. Rev. 22, 57-77. - PubMed
-
- Cerda, B. A., Horn, D. M., Breuker, K., Carpenter, B. K. & McLafferty, F. W. (1999) Eur. J. Mass Spectrom. (Chichester, England) 5, 335-338.
-
- Zubarev, R. A., Haselmann, K. F., Budnik, B., Kjeldsen, F. & Jensen, F. (2002) Eur. J. Mass Spectrom. (Chichester, England) 8, 337-349.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

