Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection
- PMID: 15213119
- PMCID: PMC427404
- DOI: 10.1128/IAI.72.7.3783-3792.2004
Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection
Abstract
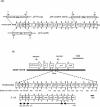
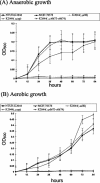
Klebsiella pneumoniae liver abscess with metastatic complications is an emerging infectious disease in Taiwan. To identify genes associated with liver infection, we used a DNA microarray to compare the transcriptional profiles of three strains causing liver abscess and three strains not associated with liver infection. There were 13 clones that showed higher RNA expression levels in the three liver infection strains, and 3 of these 13 clones contained a region that was absent in MGH 78578. Sequencing of the clones revealed the replacement of 149 bp of MGH 78578 with a 21,745-bp fragment in a liver infection strain, NTUH-K2044. This 21,745-bp fragment contained 19 open reading frames, 14 of which were proven to be associated with allantoin metabolism. The K2044 (DeltaallS) mutant showed a significant decrease of virulence in intragastric inoculation of BALB/c mice, and the prevalence of this chromosomal region was significantly higher in strains associated with liver abscess than in those that were not (19 or 32 versus 2 of 94; P = 0.0001 [chi(2) test]). Therefore, the 22-kb region may play a role in K. pneumoniae liver infection and serve as a marker for rapid identification.
Figures





References
-
- Abbott, S. 1999. Klebsiella, Enterobacter, Citrobacter, and Serratia. p. 475-482. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
-
- Benzie, I. F. F., Chung, W. Y., and B. Tomlinson. 1999. Simultaneous measurement of allantoin and urate in plasma: analytical evaluation and potential clinical application in oxidant:antioxidant balance studies. Clin. Chem. 45:901-904. - PubMed
-
- Chang, S. C., C. T. Fang, P. R. Hsueh, Y. C. Chen, and K. T. Luh. 2000. Klebsiella pneumoniae isolates causing liver abscess in Taiwan. Diagn. Microbiol. Infect. Dis. 37:279-284. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

