Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster
- PMID: 15213132
- PMCID: PMC427398
- DOI: 10.1128/IAI.72.7.3890-3901.2004
Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster
Abstract
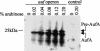
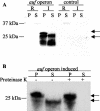
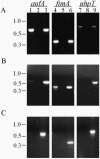
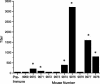
Recently, we identified a fimbrial usher gene in uropathogenic Escherichia coli strain CFT073 that is absent from an E. coli laboratory strain. Analysis of the CFT073 genome indicates that this fimbrial usher gene is part of a novel fimbrial gene cluster, aufABCDEFG. Analysis of a collection of pathogenic and commensal strains of E. coli and related species revealed that the auf gene cluster was significantly associated with uropathogenic E. coli isolates. For in vitro expression analysis of the auf gene cluster, RNA was isolated from CFT073 bacteria grown to the exponential or stationary phase in Luria-Bertani broth and reverse transcriptase PCR (RT-PCR) with oligonucleotide primers specific to the major subunit, aufA, was performed. We found that aufA is expressed in CFT073 only during the exponential growth phase; however, no expression of AufA protein was observed by Western blotting, indicating that under these conditions, the expression of the auf gene cluster is low. To determine if the auf gene cluster is expressed in vivo, RT-PCR was performed on bacteria from urine samples of mice infected with CFT073. Out of three independent experiments, we were able to detect expression of aufA at least once at 4, 24, and 48 h of infection, indicating that the auf gene cluster is expressed in the murine urinary tract. Furthermore, antisera from mice infected with CFT073 reacted with recombinant AufA in an enzyme-linked immunosorbent assay. To identify the structure encoded by the auf gene cluster, a recombinant plasmid containing the auf gene cluster under the T7 promoter was introduced into the E. coli BL-21 (AI) strain. Immunogold labeling using AufA antiserum revealed the presence of amorphous material extending from the surface of BL-21 cells. No hemagglutination or cellular adherence properties were detected in association with expression of AufA. Deletion of the entire auf gene cluster had no effect on the ability of CFT073 to colonize the kidney, bladder, or urine of mice. In addition, no significant histological differences between the parent and aufC mutant strain were observed. Therefore, Auf is a uropathogenic E. coli-associated structure that plays an uncertain role in the pathogenesis of urinary tract infections.
Figures


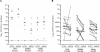

References
-
- Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type-1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. - PubMed
-
- Bahrani-Mougeot, F. K., N. W. Gunther IV, M. S. Donnenberg, and H. L. T. Mobley. 2002. Uropathogenic Escherichia coli, p. 239-268. In M. S. Donnenberg (ed.), Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press, San Diego, Calif.
-
- Bahrani-Mougeot, F. K., S. Pancholi, M. Daoust, and M. S. Donnenberg. 2001. Identification of putative urovirulence genes by subtractive cloning. J. Infect. Dis. 183(Suppl. 1):S21-S23. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

