Characterization of beta-glucan recognition site on C-type lectin, dectin 1
- PMID: 15213161
- PMCID: PMC427417
- DOI: 10.1128/IAI.72.7.4159-4171.2004
Characterization of beta-glucan recognition site on C-type lectin, dectin 1
Abstract
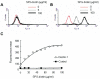
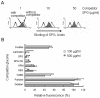
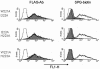
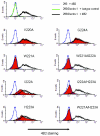
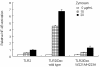
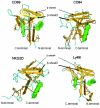
Dectin 1 is a mammalian cell surface receptor for (1-->3)-beta-d-glucans. Since (1-->3)-beta-d-glucans are commonly present on fungal cell walls, it has been suggested that dectin 1 is important for recognizing fungal invasion. In this study we tried to deduce the amino acid residues in dectin 1 responsible for beta-glucan recognition. HEK293 cells transfected with mouse dectin 1 cDNA could bind to a gel-forming (1-->3)-beta-d-glucan, schizophyllan (SPG). The binding of SPG to a dectin 1 transfectant was inhibited by pretreatment with other beta-glucans having a (1-->3)-beta-d-glucosyl linkage but not by pretreatment with alpha-glucans. Dectin 1 has a carbohydrate recognition domain (CRD) consisting of six cysteine residues that are highly conserved in C-type lectins. We prepared 32 point mutants with mutations in the CRD and analyzed their binding to SPG. Mutations at Trp(221) and His(223) resulted in decreased binding to beta-glucan. Monoclonal antibody 4B2, a dectin- 1 monoclonal antibody which had a blocking effect on the beta-glucan interaction, completely failed to bind the dectin-1 mutant W221A. A mutant with mutations in Trp(221) and His(223) did not have a collaborative effect on Toll-like receptor 2-mediated cellular activation in response to zymosan. These amino acid residues are distinct from residues in other sugar-recognizing peptide sequences of typical C-type lectins. These results suggest that the amino acid sequence W221-I222-H223 is critical for formation of a beta-glucan binding site in the CRD of dectin 1.
Figures



References
-
- Adachi, Y., N. Ohno, M. Ohsawa, K. Sato, S. Oikawa, and T. Yadomae. 1989. Physiochemical properties and antitumor activities of chemically modified derivatives of antitumor glucan “grifolan LE” from Grifola frondosa. Chem. Pharm. Bull. (Tokyo) 37:1838-1843. - PubMed
-
- Adachi, Y., N. Ohno, and T. Yadomae. 1993. Inhibitory effect of β-glucans on zymosan-mediated hydrogen peroxide production by murine peritoneal macrophages in vitro. Biol. Pharm. Bull. 16:462-467. - PubMed
-
- Adachi, Y., N. Ohno, and T. Yadomae. 1994. Preparation and antigen specificity of an anti-(1→3)-β-d-glucan antibody. Biol. Pharm. Bull. 17:1508-1512. - PubMed
-
- Anaissie, E. 1992. Opportunistic mycoses in the immunocompromised host: experience at a cancer center and review. Clin. Infect. Dis. 14:S43-S53. - PubMed
-
- Ariizumi, K., G. L. Shen, S. Shikano, S. Xu, R. Ritter III, T. Kumamoto, D. Edelbaum, A. Morita, P. R. Bergstresser, and A. Takashima. 2000. Identification of a novel, dendritic cell-associated molecule, dectin-1, by subtractive cDNA cloning. J. Biol. Chem. 275:20157-20167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

