Inter-genomic displacement via lateral gene transfer of bacterial trp operons in an overall context of vertical genealogy
- PMID: 15214963
- PMCID: PMC471576
- DOI: 10.1186/1741-7007-2-15
Inter-genomic displacement via lateral gene transfer of bacterial trp operons in an overall context of vertical genealogy
Abstract
Background: The growing conviction that lateral gene transfer plays a significant role in prokaryote genealogy opens up a need for comprehensive evaluations of gene-enzyme systems on a case-by-case basis. Genes of tryptophan biosynthesis are frequently organized as whole-pathway operons, an attribute that is expected to facilitate multi-gene transfer in a single step. We have asked whether events of lateral gene transfer are sufficient to have obscured our ability to track the vertical genealogy that underpins tryptophan biosynthesis.
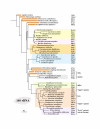
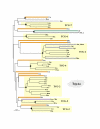
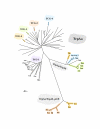
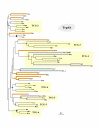
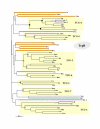
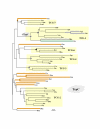
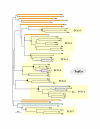
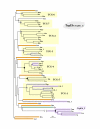
Results: In 47 complete-genome Bacteria, the genes encoding the seven catalytic domains that participate in primary tryptophan biosynthesis were distinguished from any paralogs or xenologs engaged in other specialized functions. A reliable list of orthologs with carefully ascertained functional roles has thus been assembled and should be valuable as an annotation resource. The protein domains associated with primary tryptophan biosynthesis were then concatenated, yielding single amino-acid sequence strings that represent the entire tryptophan pathway. Lateral gene transfer of several whole-pathway trp operons was demonstrated by use of phylogenetic analysis. Lateral gene transfer of partial-pathway trp operons was also shown, with newly recruited genes functioning either in primary biosynthesis (rarely) or specialized metabolism (more frequently).
Conclusions: (i) Concatenated tryptophan protein trees are congruent with 16S rRNA subtrees provided that the genomes represented are of sufficiently close phylogenetic spacing. There are currently seven tryptophan congruency groups in the Bacteria. Recognition of a succession of others can be expected in the near future, but ultimately these should coalesce to a single grouping that parallels the 16S rRNA tree (except for cases of lateral gene transfer). (ii) The vertical trace of evolution for tryptophan biosynthesis can be deduced. The daunting complexities engendered by paralogy, xenology, and idiosyncrasies of nomenclature at this point in time have necessitated an expert-assisted manual effort to achieve a correct analysis. Once recognized and sorted out, paralogy and xenology can be viewed as features that enrich evolutionary histories.
Figures




References
-
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rate of change and exchange. J Mol Evol. 1997;44:383–397. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

