Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae
- PMID: 15215087
- PMCID: PMC434173
- DOI: 10.1128/AAC.48.7.2400-2408.2004
Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae
Abstract
We investigated the diversity of the chromosomal class A beta-lactamase gene in Klebsiella pneumoniae in order to study the evolution of the gene. A 789-bp portion was sequenced in a panel of 28 strains, representative of three phylogenetic groups, KpI, KpII, and KpIII, recently identified in K. pneumoniae and of different chromosomal beta-lactamase variants previously identified. Three groups of sequences were found, two of them corresponding to the families SHV (pI 7.6) and LEN (pI 7.1), respectively, and one, more heterogeneous, corresponding to a new family that we named OKP (for other K. pneumoniae beta-lactamase). Levels of susceptibility to ampicillin, cefuroxime, cefotaxime, ceftazidime, and aztreonam and inhibition by clavulanic acid were similar in the three groups. One new SHV variant, seven new LEN variants, and four OKP variants were identified. The OKP variants formed two subgroups based on nucleotide sequences, one with pIs of 7.8 and 8.1 and the other with pIs of 6.5 and 7.0. The nucleotide sequences of the housekeeping genes gyrA, coding for subunit A of gyrase, and mdh, coding for malate dehydrogenase, were also determined. Phylogenetic analysis of the three genes studied revealed parallel evolution, with the SHV, OKP, and LEN beta-lactamase families corresponding to the phylogenetic groups KpI, KpII, and KpIII, respectively. This correspondence was fully confirmed for 34 additional strains in PCR assays specific for the three beta-lactamase families. We estimated the time since divergence of the phylogenetic groups KpI and KpIII at between 6 and 28 million years, confirming the ancient presence of the beta-lactamase gene in the genome of K. pneumoniae.
Figures
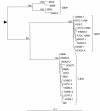
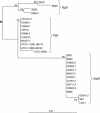
References
-
- Ambler, R. P. 1980. The structure of beta-lactamases. Phil. Trans. R. Soc. Lond. B 289:321-331. - PubMed
-
- Arakawa, Y., M. Ohta, N. Kido, Y. Fujii, T. Komatsu, and N. Kato. 1986. Close evolutionary relationship between the chromosomally encoded beta-lactamase gene of Klebsiella pneumoniae and the TEM beta-lactamase gene mediated by R plasmids. FEBS Lett. 207:69-74. - PubMed
-
- Barlow, M., and B. G. Hall. 2002. Phylogenetic analysis shows that the OXA beta-lactamase genes have been on plasmids for millions of years. J. Mol. Evol. 55:314-321. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

