Saccharomyces cerevisiae multidrug transporter Qdr2p (Yil121wp): localization and function as a quinidine resistance determinant
- PMID: 15215105
- PMCID: PMC434225
- DOI: 10.1128/AAC.48.7.2531-2537.2004
Saccharomyces cerevisiae multidrug transporter Qdr2p (Yil121wp): localization and function as a quinidine resistance determinant
Abstract
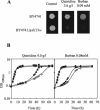
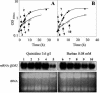
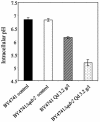
This work reports the functional analysis of Saccharomyces cerevisiae open reading frame YIL121w, encoding a member of a family of drug:H(+) antiporters with 12 predicted membrane-spanning segments (DHA12 family). Like its close homologue Qdr1p, Yil121wp was localized at the plasma membrane, and its increased expression also led to increased tolerance to the antiarrhythmia and antimalarial drug quinidine. The quinidine resistance phenotype was confirmed for different yeast strains and growth media, including a prototrophic strain, and YIL121w was named the QDR2 gene. Both QDR1 and QDR2 were also implicated in yeast resistance to the herbicide barban (4-chloro-2-butynyl [3-chlorophenyl] carbamate), and the genes are functionally interchangeable with respect to both resistance phenotypes. The average intracellular pH of a yeast population challenged with quinidine added to the acidic growth medium was significantly below the intracellular pH of the unstressed population, suggesting plasma membrane permeabilization by quinidine with consequent increase of the H(+) influx rate. For the same extracellular quinidine concentration, internal acidification was more intense for the Deltaqdr2 deletant compared with the parental strain. Although QDR2 transcription was not enhanced in response to quinidine, the results confirmed that Qdr2p is involved in the active export of quinidine out of the cell, thus conferring resistance to the drug.
Figures
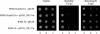



Similar articles
-
Resistance and adaptation to quinidine in Saccharomyces cerevisiae: role of QDR1 (YIL120w), encoding a plasma membrane transporter of the major facilitator superfamily required for multidrug resistance.Antimicrob Agents Chemother. 2001 May;45(5):1528-34. doi: 10.1128/AAC.45.5.1528-1534.2001. Antimicrob Agents Chemother. 2001. PMID: 11302822 Free PMC article.
-
Saccharomyces cerevisiae multidrug resistance transporter Qdr2 is implicated in potassium uptake, providing a physiological advantage to quinidine-stressed cells.Eukaryot Cell. 2007 Feb;6(2):134-42. doi: 10.1128/EC.00290-06. Epub 2006 Dec 22. Eukaryot Cell. 2007. PMID: 17189489 Free PMC article.
-
The yeast multidrug transporter Qdr3 (Ybr043c): localization and role as a determinant of resistance to quinidine, barban, cisplatin, and bleomycin.Biochem Biophys Res Commun. 2005 Feb 18;327(3):952-9. doi: 10.1016/j.bbrc.2004.12.097. Biochem Biophys Res Commun. 2005. PMID: 15649438
-
Functional analysis of fungal drug efflux transporters by heterologous expression in Saccharomyces cerevisiae.Jpn J Infect Dis. 2005 Feb;58(1):1-7. Jpn J Infect Dis. 2005. PMID: 15728981 Review.
-
The multidrug resistance transporters of the major facilitator superfamily, 6 years after disclosure of Saccharomyces cerevisiae genome sequence.J Biotechnol. 2002 Sep 25;98(2-3):215-26. doi: 10.1016/s0168-1656(02)00133-5. J Biotechnol. 2002. PMID: 12141988 Review.
Cited by
-
Host-Pathogen Interactions Mediated by MDR Transporters in Fungi: As Pleiotropic as it Gets!Genes (Basel). 2018 Jul 2;9(7):332. doi: 10.3390/genes9070332. Genes (Basel). 2018. PMID: 30004464 Free PMC article. Review.
-
Yeast toxicogenomics: genome-wide responses to chemical stresses with impact in environmental health, pharmacology, and biotechnology.Front Genet. 2012 Apr 19;3:63. doi: 10.3389/fgene.2012.00063. eCollection 2012. Front Genet. 2012. PMID: 22529852 Free PMC article.
-
Transcriptomic profiling of the Saccharomyces cerevisiae response to quinine reveals a glucose limitation response attributable to drug-induced inhibition of glucose uptake.Antimicrob Agents Chemother. 2009 Dec;53(12):5213-23. doi: 10.1128/AAC.00794-09. Epub 2009 Oct 5. Antimicrob Agents Chemother. 2009. PMID: 19805573 Free PMC article.
-
The antifungal plant defensin AtPDF2.3 from Arabidopsis thaliana blocks potassium channels.Sci Rep. 2016 Aug 30;6:32121. doi: 10.1038/srep32121. Sci Rep. 2016. PMID: 27573545 Free PMC article.
-
The C-terminus of the cargo receptor Erv14 affects COPII vesicle formation and cargo delivery.J Cell Sci. 2023 Feb 1;136(3):jcs260527. doi: 10.1242/jcs.260527. Epub 2023 Feb 6. J Cell Sci. 2023. PMID: 36651113 Free PMC article.
References
-
- Bauer, B. E., D. Rossington, M. Mollapour, Y. Mamnun, K. Kuchler, and P. W. Piper. 2003. Weak organic acid stress inhibits aromatic amino acid uptake in yeast, causing a strong influence of amino acid auxotrophies in phenotypes of membrane transporter mutants. Eur. J. Biochem. 270:3189-3195. - PubMed
-
- Brôco, N., S. Tenreiro, C. A. Viegas, and I. Sá-Correia. 1999. FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance in Saccharomyces cerevisiae and its benomyl-induced expression is dependent on Pdr3 transcriptional regulator. Yeast 15:1595-1608. - PubMed
-
- do Valle Matta, M. A., J. L. Jonniaux, E. Balzi, A. Goffeau, and B. van den Hazel. 2001. Novel target genes of the yeast regulator Pdr1p: a contribution of the TPO1 gene in resistance to quinidine and other drugs. Gene. 272:111-119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

