Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC
- PMID: 15215121
- PMCID: PMC434217
- DOI: 10.1128/AAC.48.7.2637-2651.2004
Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC
Abstract
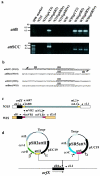
Staphylococcal cassette chromosome mec (SCCmec) is a mobile genetic element composed of the mec gene complex, which encodes methicillin resistance, and the ccr gene complex, which encodes the recombinases responsible for its mobility. The mec gene complex has been classified into four classes, and the ccr gene complex has been classified into three allotypes. Different combinations of mec gene complex classes and ccr gene complex types have so far defined four types of SCCmec elements. Now we introduce the fifth allotype of SCCmec, which was found on the chromosome of a community-acquired methicillin-resistant Staphylococcus aureus strain (strain WIS [WBG8318]) isolated in Australia. The element shared the same chromosomal integration site with the four extant types of SCCmec and the characteristic nucleotide sequences at the chromosome-SCCmec junction regions. The novel SCCmec carried mecA bracketed by IS431 (IS431-mecA-DeltamecR1-IS431), which is designated the class C2 mec gene complex; and instead of ccrA and ccrB genes, it carried a single copy of a gene homologue that encoded cassette chromosome recombinase. Since the open reading frame (ORF) was found to encode an enzyme which catalyzes the precise excision as well as site- and orientation-specific integration of the element, we designated the ORF cassette chromosome recombinase C (ccrC), and we designated the element type V SCCmec. Type V SCCmec is a small SCCmec element (28 kb) and does not carry any antibiotic resistance genes besides mecA. Unlike the extant SCCmec types, it carries a set of foreign genes encoding a restriction-modification system that might play a role in the stabilization of the element on the chromosome.
Figures





References
-
- Ayliffe, G. A. 1997. The progressive international spread of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 24:74-79. - PubMed
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. - PubMed
-
- Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA 282:1123-1125. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

