Complete release of (5'S)-8,5'-cyclo-2'-deoxyadenosine from dinucleotides, oligodeoxynucleotides and DNA, and direct comparison of its levels in cellular DNA with other oxidatively induced DNA lesions
- PMID: 15215337
- PMCID: PMC443555
- DOI: 10.1093/nar/gnh087
Complete release of (5'S)-8,5'-cyclo-2'-deoxyadenosine from dinucleotides, oligodeoxynucleotides and DNA, and direct comparison of its levels in cellular DNA with other oxidatively induced DNA lesions
Abstract
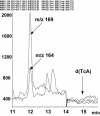
8,5'-cyclopurine-2'-deoxynucleosides in DNA are repaired by nucleotide-excision repair, and act as strong blocks to DNA polymerases, RNA polymerase II and transcription factor binding. Thus, it is important to accurately determine the level of these lesions in DNA. There is controversy in the literature regarding the ability of different enzymes to release these compounds from oligodeoxynucleotides or DNA. We used liquid chromatography/mass spectrometry (LC/MS) to investigate the ability of several enzymes to release (5'S)-8,5'-cyclo-2'-deoxyadenosine [(5'S)-cdA] from dinucleotides and oligodeoxynucleotides and from DNA. The data show that (5'S)-cdA is completely released from DNA by hydrolysis with nuclease P1, snake venom phosphodiesterase and alkaline phosphatase. The identity of the normal nucleoside 5' to the (5'S)-cdA had a significant effect on its release. Using LC/MS, we also showed that the levels of (5'S)-cdA were within an order of magnitude of those of 8-hydroxy-2'-deoxyguanosine, and three times higher than those of 8-hydroxy-2'-deoxyadenosine in pig liver DNA. Different DNA isolation methods affected the levels of the latter two lesions, but did not influence those of (5'S)-cdA. We conclude that (5'S)-cdA can be completely released from DNA by enzymic hydrolysis, and the level of (5'S)-cdA in tissue DNA is comparable to those of other oxidatively induced DNA lesions.
Figures

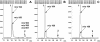
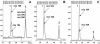
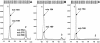
References
-
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. - PubMed
-
- Dizdaroglu M. (1992) Oxidative damage to DNA in mammalian chromatin. Mutat. Res., 275, 331–342. - PubMed
-
- Breen A.P. and Murphy,J.A. (1995) Reactions of oxyl radicals with DNA. Free Radic. Biol. Med., 18, 1033–1077. - PubMed
-
- Wallace S.S. (1998) Enzymatic processing of radiation-induced free radical damage in DNA. Radiat. Res., 150, S60–S79. - PubMed
-
- Dizdaroglu M., Jaruga,P., Birincioglu,M. and Rodriguez,H. (2002) Free radical-induced damage to DNA: mechanisms and measurement. Free Radic. Biol. Med., 32, 1102–1115. - PubMed

