Riboswitch finder--a tool for identification of riboswitch RNAs
- PMID: 15215370
- PMCID: PMC441490
- DOI: 10.1093/nar/gkh352
Riboswitch finder--a tool for identification of riboswitch RNAs
Abstract
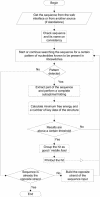
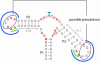
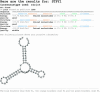
We describe a dedicated RNA motif search program and web server to identify RNA riboswitches. The Riboswitch finder analyses a given sequence using the web interface, checks specific sequence elements and secondary structure, calculates and displays the energy folding of the RNA structure and runs a number of tests including this information to determine whether high-sensitivity riboswitch motifs (or variants) according to the Bacillus subtilis type are present in the given RNA sequence. Batch-mode determination (all sequences input at once and separated by FASTA format) is also possible. The program has been implemented and is available both as local software for in-house installation and as a web server at http://www.biozentrum.uni-wuerzburg.de/bioinformatik/Riboswitch/.
Figures

References
-
- Stormo G.D. (2003) New tricks for an old dogma: riboswitches as cis-only regulatory systems. Mol. Cell, 11, 1419–1420. - PubMed
-
- Mandal M., Boese,B., Barrick,J.E., Winkler,W.C. and Breaker,R.R. (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell, 113, 577–586. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

