Proteome Analyst: custom predictions with explanations in a web-based tool for high-throughput proteome annotations
- PMID: 15215412
- PMCID: PMC441623
- DOI: 10.1093/nar/gkh485
Proteome Analyst: custom predictions with explanations in a web-based tool for high-throughput proteome annotations
Abstract
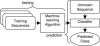
Proteome Analyst (PA) (http://www.cs.ualberta.ca/~bioinfo/PA/) is a publicly available, high-throughput, web-based system for predicting various properties of each protein in an entire proteome. Using machine-learned classifiers, PA can predict, for example, the GeneQuiz general function and Gene Ontology (GO) molecular function of a protein. In addition, PA is currently the most accurate and most comprehensive system for predicting subcellular localization, the location within a cell where a protein performs its main function. Two other capabilities of PA are notable. First, PA can create a custom classifier to predict a new property, without requiring any programming, based on labeled training data (i.e. a set of examples, each with the correct classification label) provided by a user. PA has been used to create custom classifiers for potassium-ion channel proteins and other general function ontologies. Second, PA provides a sophisticated explanation feature that shows why one prediction is chosen over another. The PA system produces a Naïve Bayes classifier, which is amenable to a graphical and interactive approach to explanations for its predictions; transparent predictions increase the user's confidence in, and understanding of, PA.
Figures





References
-
- Andrade M.A., Brown,N.P., Leroy,C., Hoersch,S., de Daruvar,A., Reich,C., Franchini,A., Tamames,J., Valencia,A., Ouzounis,C. and Sander,C. (1999) Automated genome sequence analysis and annotation. Bioinformatics, 15, 391–412. - PubMed
-
- Kitson D.H., Badretdinov,A., Zhu,Z.Y., Velikanov,M., Edwards,D.J., Olszewski,K., Szalma,S. and Yan,L. (2002) Functional annotation of proteomic sequences based on consensus of sequence and structural analysis. Brief. Bioinformatics, 3, 32–44. - PubMed
-
- Frishman D., Albermann,K., Hani,J., Heumann,K., Metanomski,A., Zollner,A. and Mewes,H.W. (2001) Functional and structural genomics using PEDANT. Bioinformatics, 17, 44–57. - PubMed

