MITOPRED: a web server for the prediction of mitochondrial proteins
- PMID: 15215413
- PMCID: PMC441512
- DOI: 10.1093/nar/gkh374
MITOPRED: a web server for the prediction of mitochondrial proteins
Abstract
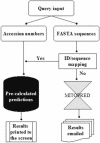
MITOPRED web server enables prediction of nucleus-encoded mitochondrial proteins in all eukaryotic species. Predictions are made using a new algorithm based primarily on Pfam domain occurrence patterns in mitochondrial and non-mitochondrial locations. Pre-calculated predictions are instantly accessible for proteomes of Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila, Homo sapiens, Mus musculus and Arabidopsis species as well as all the eukaryotic sequences in the Swiss-Prot and TrEMBL databases. Queries, at different confidence levels, can be made through four distinct options: (i) entering Swiss-Prot/TrEMBL accession numbers; (ii) uploading a local file with such accession numbers; (iii) entering protein sequences; (iv) uploading a local file containing protein sequences in FASTA format. Automated updates are scheduled for the pre-calculated prediction database so as to provide access to the most current data. The server, its documentation and the data are available from http://mitopred.sdsc.edu.
Figures
References
-
- Huh W.-K., Falvo,J.V., Gerke,L.C., Carroll,A.S., Howson,R.W., Weissman,J.S. and O'Shea,E.K. (2003) Global analysis of protein localization in budding yeast. Nature, 425, 686–691. - PubMed
-
- Nielsen H., Engelbrech,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. - PubMed
-
- Fujiwara Y., Asogawa,M. and Nakai,K. (1997) Prediction of mitochondrial targeting signals using hidden Markov models. In Miyano,S. and Takagi,T. (eds), Genome Informatics. Universal Academy Press, Inc., Tokyo, Japan, pp. 53–60. - PubMed
-
- Nakai K. and Horton,P. (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem., Sci., 24, 34–36. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases