PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins
- PMID: 15215419
- PMCID: PMC441555
- DOI: 10.1093/nar/gkh417
PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins
Abstract
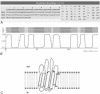
The beta-barrel outer membrane proteins constitute one of the two known structural classes of membrane proteins. Whereas there are several different web-based predictors for alpha-helical membrane proteins, currently there is no freely available prediction method for beta-barrel membrane proteins, at least with an acceptable level of accuracy. We present here a web server (PRED-TMBB, http://bioinformatics.biol.uoa.gr/PRED-TMBB) which is capable of predicting the transmembrane strands and the topology of beta-barrel outer membrane proteins of Gram-negative bacteria. The method is based on a Hidden Markov Model, trained according to the Conditional Maximum Likelihood criterion. The model was retrained and the training set now includes 16 non-homologous outer membrane proteins with structures known at atomic resolution. The user may submit one sequence at a time and has the option of choosing between three different decoding methods. The server reports the predicted topology of a given protein, a score indicating the probability of the protein being an outer membrane beta-barrel protein, posterior probabilities for the transmembrane strand prediction and a graphical representation of the assumed position of the transmembrane strands with respect to the lipid bilayer.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

