PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations
- PMID: 15215472
- PMCID: PMC441519
- DOI: 10.1093/nar/gkh381
PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations
Abstract
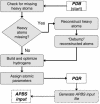
Continuum solvation models, such as Poisson-Boltzmann and Generalized Born methods, have become increasingly popular tools for investigating the influence of electrostatics on biomolecular structure, energetics and dynamics. However, the use of such methods requires accurate and complete structural data as well as force field parameters such as atomic charges and radii. Unfortunately, the limiting step in continuum electrostatics calculations is often the addition of missing atomic coordinates to molecular structures from the Protein Data Bank and the assignment of parameters to biomolecular structures. To address this problem, we have developed the PDB2PQR web service (http://agave.wustl.edu/pdb2pqr/). This server automates many of the common tasks of preparing structures for continuum electrostatics calculations, including adding a limited number of missing heavy atoms to biomolecular structures, estimating titration states and protonating biomolecules in a manner consistent with favorable hydrogen bonding, assigning charge and radius parameters from a variety of force fields, and finally generating 'PQR' output compatible with several popular computational biology packages. This service is intended to facilitate the setup and execution of electrostatics calculations for both experts and non-experts and thereby broaden the accessibility to the biological community of continuum electrostatics analyses of biomolecular systems.
Figures
References
-
- Baker N.A. and McCammon,J.A. (2002) Electrostatic interactions. In Bourne,P. and Weissig,H. (eds.), Structural Bioinformatics. John Wiley & Sons, Inc., New York.
-
- Gilson M. (2000) Introduction to continuum electrostatics. In Beard,D.A. (ed.), Biophysics Textbook Online. Biophysical Society, Bethesda, MD, Vol. Computational Biology.
-
- Darden T.A. (2001) Treatment of long-range forces and potential. In Becker,O.M., MacKerell,A.D.J., Roux,B. and Watanabe,M. (eds), Computational Biochemistry and Biophysics. Marcel Dekker, Inc., New York, pp. 91–114.
-
- Roux B. (2001) Implicit solvent models. In Becker,O.M., MacKerell,A.D.J., Roux,B. and Watanabe,M. (eds), Computational Biochemistry and Biophysics. Marcel Dekker, New York, pp. 133–152.
-
- Davis M.E. and McCammon,J.A. (1990) Electrostatics in biomolecular structure and dynamics. Chem. Rev., 94, 7684–7692.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources