Analysis of the "thermodynamic information content" of a Homo sapiens structural database reveals hierarchical thermodynamic organization
- PMID: 15215522
- PMCID: PMC2279918
- DOI: 10.1110/ps.04706204
Analysis of the "thermodynamic information content" of a Homo sapiens structural database reveals hierarchical thermodynamic organization
Abstract
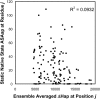
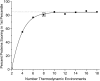
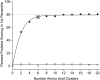
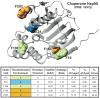
Classification of the amounts and types of lower order structural elements in proteins is a prerequisite to effective comparisons between protein folds. In an effort to provide an additional vehicle for fold comparison, we present an alternative classification scheme whereby protein folds are represented in statistical thermodynamic terms in such a way as to illuminate the energetic building blocks within protein structures. The thermodynamic relationship is examined between amino acid sequences and the conformational ensembles for a database of 159 Homo sapiens protein structures ranging from 50 to 250 amino acids. Using hierarchical clustering, it is shown through fold-recognition experiments that (1) eight thermodynamic environmental descriptors sufficiently accounts for the energetic variation within the native state ensembles of the H. sapiens structural database, (2) an amino acid library of only six residue types is sufficient to encode >90% of the thermodynamic information required for fold specificity in the entire database, and (3) structural resolution of the statistically derived environments reveals sequential cooperative segments throughout the protein, which are independent of secondary structure. As the first level of thermodynamic organization in proteins, these segments represent the thermodynamic counterpart to secondary structure.
Figures




Similar articles
-
Denatured-state energy landscapes of a protein structural database reveal the energetic determinants of a framework model for folding.J Mol Biol. 2008 Sep 19;381(5):1184-201. doi: 10.1016/j.jmb.2008.06.046. Epub 2008 Jun 24. J Mol Biol. 2008. PMID: 18616947 Free PMC article.
-
Thermodynamic propensities of amino acids in the native state ensemble: implications for fold recognition.Protein Sci. 2001 May;10(5):1032-45. doi: 10.1110/ps.01601. Protein Sci. 2001. PMID: 11316884 Free PMC article.
-
Thermodynamic environments in proteins: fundamental determinants of fold specificity.Protein Sci. 2002 Aug;11(8):1945-57. doi: 10.1110/ps.0203202. Protein Sci. 2002. PMID: 12142449 Free PMC article.
-
Conformational dynamics and ensembles in protein folding.Annu Rev Biophys Biomol Struct. 2007;36:395-412. doi: 10.1146/annurev.biophys.36.040306.132608. Annu Rev Biophys Biomol Struct. 2007. PMID: 17291180 Review.
-
[Thermodynamic databases for proteins and interactions].Tanpakushitsu Kakusan Koso. 2002 Jun;47(8 Suppl):1071-5. Tanpakushitsu Kakusan Koso. 2002. PMID: 12099025 Review. Japanese. No abstract available.
Cited by
-
Investigating homology between proteins using energetic profiles.PLoS Comput Biol. 2010 Mar 26;6(3):e1000722. doi: 10.1371/journal.pcbi.1000722. PLoS Comput Biol. 2010. PMID: 20361049 Free PMC article.
-
Denatured-state energy landscapes of a protein structural database reveal the energetic determinants of a framework model for folding.J Mol Biol. 2008 Sep 19;381(5):1184-201. doi: 10.1016/j.jmb.2008.06.046. Epub 2008 Jun 24. J Mol Biol. 2008. PMID: 18616947 Free PMC article.
-
Hidden dynamic signatures drive substrate selectivity in the disordered phosphoproteome.Proc Natl Acad Sci U S A. 2020 Sep 22;117(38):23606-23616. doi: 10.1073/pnas.1921473117. Epub 2020 Sep 8. Proc Natl Acad Sci U S A. 2020. PMID: 32900925 Free PMC article.
-
Temperature and urea have opposing impacts on polyproline II conformational bias.Biochemistry. 2013 Feb 5;52(5):949-58. doi: 10.1021/bi301435p. Epub 2013 Jan 27. Biochemistry. 2013. PMID: 23350874 Free PMC article.
-
Predicting the energetics of conformational fluctuations in proteins from sequence: a strategy for profiling the proteome.Structure. 2008 Nov 12;16(11):1627-37. doi: 10.1016/j.str.2008.08.016. Structure. 2008. PMID: 19000815 Free PMC article.
References
-
- Anfinsen, C.B. 1973. Principles that govern the folding of protein chains. Science 181 223–230. - PubMed
-
- Babu, C.R., Hilser, V.J., and Wand, A.J. 2004. Direct access to the cooperative substructure of proteins and the protein ensemble via cold denaturation. Nat. Struct. Mol. Biol. 11 352–357. - PubMed
-
- Bai, Y. and Englander, S.W. 1996. Future directions in folding: The multi-state nature of protein structure. Proteins 24 145–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

