Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2
- PMID: 15215894
- PMCID: PMC514941
- DOI: 10.1038/sj.emboj.7600274
Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2
Abstract
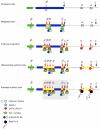
How does an emerging transcriptional programme regulate individual genes as stem cells undergo lineage commitment, differentiation and maturation? To answer this, we have analysed the dynamic protein/DNA interactions across 130 kb of chromatin containing the mouse alpha-globin cluster in cells representing all stages of differentiation from stem cells to mature erythroblasts. The alpha-gene cluster appears to be inert in pluripotent cells, but priming of expression begins in multipotent haemopoietic progenitors via GATA-2. In committed erythroid progenitors, GATA-2 is replaced by GATA-1 and binding is extended to additional sites including the alpha-globin promoters. Both GATA-1 and GATA-2 nucleate the binding of various protein complexes including SCL/LMO2/E2A/Ldb-1 and NF-E2. Changes in protein/DNA binding are accompanied by sequential alterations in long-range histone acetylation and methylation. The recruitment of polymerase II, which ultimately leads to a rapid increase in alpha-globin transcription, occurs late in maturation. These studies provide detailed evidence for the more general hypothesis that commitment and differentiation are primarily driven by the sequential appearance of key transcriptional factors, which bind chromatin at specific, high-affinity sites.
Figures









References
-
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH (1993a) Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362: 722–728 - PubMed
-
- Anguita E, Sharpe JA, Sloane-Stanley JA, Tufarelli C, Higgs DR, Wood WG (2002) Deletion of the mouse alpha-globin regulatory element (HS-26) has an unexpectedly mild phenotype. Blood 100: 3450–3456 - PubMed
-
- Bottardi S, Aumont A, Grosveld F, Milot E (2003) Developmental stage-specific epigenetic control of human beta-globin gene expression is potentiated in hematopoietic progenitor cells prior to their transcriptional activation. Blood 102: 3989–3997 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

