Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF
- PMID: 15215896
- PMCID: PMC514944
- DOI: 10.1038/sj.emboj.7600280
Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF
Abstract
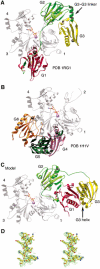
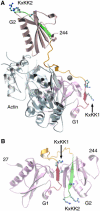
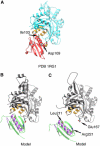
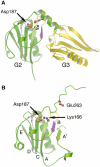
The actin filament-severing functionality of gelsolin resides in its N-terminal three domains (G1-G3). We have determined the structure of this fragment in complex with an actin monomer. The structure reveals the dramatic domain rearrangements that activate G1-G3, which include the replacement of interdomain interactions observed in the inactive, calcium-free protein by new contacts to actin, and by a novel G2-G3 interface. Together, these conformational changes are critical for actin filament severing, and we suggest that their absence leads to the disease Finnish-type familial amyloidosis. Furthermore, we propose that association with actin drives the calcium-independent activation of isolated G1-G3 during apoptosis, and that a similar mechanism operates to activate native gelsolin at micromolar levels of calcium. This is the first structure of a filament-binding protein bound to actin and it sets stringent, high-resolution limitations on the arrangement of actin protomers within the filament.
Figures


References
-
- Allen PG (2003) Actin filament uncapping localizes to ruffling lamellae and rocketing vesicles. Nat Cell Biol 5: 972–979 - PubMed
-
- Azuma T, Koths K, Flanagan L, Kwiatkowski D (2000) Gelsolin in complex with phosphatidylinositol 4,5-bisphosphate inhibits caspase-3 and -9 to retard apoptotic progression. J Biol Chem 275: 3761–3766 - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Crosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 - PubMed
-
- Burtnick LD, Koepf EK, Grimes J, Jones EY, Stuart DI, McLaughlin PJ, Robinson RC (1997) The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation. Cell 90: 661–670 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

