Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions
- PMID: 15220348
- PMCID: PMC2757098
- DOI: 10.1074/jbc.M406225200
Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions
Abstract
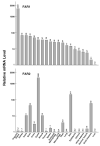
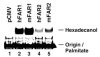
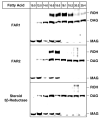
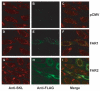
The conversion of fatty acids to fatty alcohols is required for the synthesis of wax monoesters and ether lipids. The mammalian enzymes that synthesize fatty alcohols have not been identified. Here, an in silico approach was used to discern two putative reductase enzymes designated FAR1 and FAR2. Expression studies in intact cells showed that FAR1 and FAR2 cDNAs encoded isozymes that reduced fatty acids to fatty alcohols. Fatty acyl-CoA esters were the substrate of FAR1, and the enzyme required NADPH as a cofactor. FAR1 preferred saturated and unsaturated fatty acids of 16 or 18 carbons as substrates, whereas FAR2 preferred saturated fatty acids of 16 or 18 carbons. Confocal light microscopy indicated that FAR1 and FAR2 were localized in the peroxisome. The FAR1 mRNA was detected in many mouse tissues with the highest level found in the preputial gland, a modified sebaceous gland. The FAR2 mRNA was more restricted in distribution and most abundant in the eyelid, which contains wax-laden meibomian glands. Both FAR mRNAs were present in the brain, a tissue rich in ether lipids. The data suggest that fatty alcohol synthesis in mammals is accomplished by two fatty acyl-CoA reductase isozymes that are expressed at high levels in tissues known to synthesize wax monoesters and ether lipids.
Figures



References
-
- Downing DT, Strauss JS. J. Investig. Dermatol. 1974;62:228–244. - PubMed
-
- Nikkari T. J. Investig. Dermatol. 1974;62:257–267. - PubMed
-
- Jenks MA, Eigenbrode SD, Lemieux B. In: The Arabidopsis Book. Somerville CR, Meyerowitz EM, editors. American Society of Plant Biologists; Rockville, MD: 2002. doi/10.1199/tab.0016. http://www.aspb.org/publications/arabidopsis/
-
- Kunst L, Samuels AL. Prog. Lipid Res. 2003;42:51–80. - PubMed
-
- Wykle RL, Malone B, Snyder F. J. Lipid Res. 1979;20:890–896. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

