Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes
- PMID: 15220413
- PMCID: PMC434129
- DOI: 10.1128/JVI.78.14.7400-7409.2004
Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes
Abstract
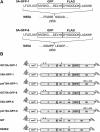
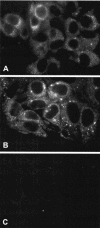
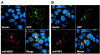
Hepatitis C virus (HCV) replicates its genome in a membrane-associated replication complex, composed of viral proteins, replicating RNA and altered cellular membranes. We describe here HCV replicons that allow the direct visualization of functional HCV replication complexes. Viable replicons selected from a library of Tn7-mediated random insertions in the coding sequence of nonstructural protein 5A (NS5A) allowed the identification of two sites near the NS5A C terminus that tolerated insertion of heterologous sequences. Replicons encoding green fluorescent protein (GFP) at these locations were only moderately impaired for HCV RNA replication. Expression of the NS5A-GFP fusion protein could be demonstrated by immunoblot, indicating that the GFP was retained during RNA replication and did not interfere with HCV polyprotein processing. More importantly, expression levels were robust enough to allow direct visualization of the fusion protein by fluorescence microscopy. NS5A-GFP appeared as brightly fluorescing dot-like structures in the cytoplasm. By confocal laser scanning microscopy, NS5A-GFP colocalized with other HCV nonstructural proteins and nascent viral RNA, indicating that the dot-like structures, identified as membranous webs by electron microscopy, represent functional HCV replication complexes. These findings reveal an unexpected flexibility of the C-terminal domain of NS5A and provide tools for studying the formation and turnover of HCV replication complexes in living cells.
Figures




Similar articles
-
Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA.J Gen Virol. 2007 Feb;88(Pt 2):470-475. doi: 10.1099/vir.0.82363-0. J Gen Virol. 2007. PMID: 17251564
-
Genetic complementation of hepatitis C virus nonstructural protein functions associated with replication exhibits requirements that differ from those for virion assembly.J Virol. 2014 Mar;88(5):2748-62. doi: 10.1128/JVI.03588-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352463 Free PMC article.
-
Tagging of NS5A expressed from a functional hepatitis C virus replicon.J Gen Virol. 2006 Mar;87(Pt 3):635-640. doi: 10.1099/vir.0.81553-0. J Gen Virol. 2006. PMID: 16476985
-
Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication?Virology. 2007 Jul 20;364(1):1-9. doi: 10.1016/j.virol.2007.01.042. Epub 2007 Apr 2. Virology. 2007. PMID: 17400273 Review.
-
Hepatitis C virus NS5A: tales of a promiscuous protein.J Gen Virol. 2004 Sep;85(Pt 9):2485-2502. doi: 10.1099/vir.0.80204-0. J Gen Virol. 2004. PMID: 15302943 Review.
Cited by
-
Reverse Genetic Approaches for the Generation of Full Length and Subgenomic Replicon of EV71 Virus.Front Microbiol. 2021 May 20;12:665879. doi: 10.3389/fmicb.2021.665879. eCollection 2021. Front Microbiol. 2021. PMID: 34093481 Free PMC article.
-
Hepatitis C Virus Is Released via a Noncanonical Secretory Route.J Virol. 2016 Nov 14;90(23):10558-10573. doi: 10.1128/JVI.01615-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27630244 Free PMC article.
-
Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes.J Virol. 2007 May;81(9):4591-603. doi: 10.1128/JVI.02144-06. Epub 2007 Feb 14. J Virol. 2007. PMID: 17301154 Free PMC article.
-
Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA.J Virol. 2005 Feb;79(4):2346-55. doi: 10.1128/JVI.79.4.2346-2355.2005. J Virol. 2005. PMID: 15681435 Free PMC article.
-
Insertion of Exogenous Genes within the ORF1a Coding Region of Porcine Astrovirus.Viruses. 2021 Oct 21;13(11):2119. doi: 10.3390/v13112119. Viruses. 2021. PMID: 34834925 Free PMC article.
References
-
- Bartenschlager, R. 2002. Hepatitis C virus replicons: potential for drug development. Nat. Rev. Drug Discov. 1:911-916. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

