Human immunodeficiency virus type 1 evades T-helper responses by exploiting antibodies that suppress antigen processing
- PMID: 15220439
- PMCID: PMC434093
- DOI: 10.1128/JVI.78.14.7645-7652.2004
Human immunodeficiency virus type 1 evades T-helper responses by exploiting antibodies that suppress antigen processing
Abstract
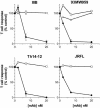
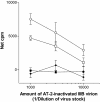
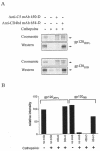
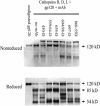
T-helper responses are important for controlling chronic viral infections, yet T-helper responses specific to human immunodeficiency virus type 1 (HIV-1), particularly to envelope glycoproteins, are lacking in the vast majority of HIV-infected individuals. It was previously shown that the presence of antibodies to the CD4-binding domain (CD4bd) of HIV-1 glycoprotein 120 (gp120) prevents T-helper responses to gp120, but their suppressive mechanisms were undefined (C. E. Hioe et al., J. Virol. 75:10950-10957, 2001). The present study demonstrates that gp120, when complexed to anti-CD4bd antibodies, becomes more resistant to proteolysis by lysosomal enzymes from antigen-presenting cells such that peptide epitopes are not released and presented efficiently by major histocompatibility complex class II molecules to gp120-specific CD4 T cells. Antibodies to other gp120 regions do not confer this effect. Thus, HIV may evade anti-viral T-helper responses by inducing and exploiting antibodies that conceal the virus envelope antigens from T cells.
Figures

Similar articles
-
Inhibition of human immunodeficiency virus type 1 gp120 presentation to CD4 T cells by antibodies specific for the CD4 binding domain of gp120.J Virol. 2001 Nov;75(22):10950-7. doi: 10.1128/JVI.75.22.10950-10957.2001. J Virol. 2001. PMID: 11602735 Free PMC article.
-
Anti-CD4-binding domain antibodies complexed with HIV type 1 glycoprotein 120 inhibit CD4+ T cell-proliferative responses to glycoprotein 120.AIDS Res Hum Retroviruses. 2000 Jun 10;16(9):893-905. doi: 10.1089/08892220050042837. AIDS Res Hum Retroviruses. 2000. PMID: 10875615
-
HIV-1-infected patients with envelope-specific lymphoproliferation or long-term nonprogression lack antibodies suppressing glycoprotein 120 antigen presentation.J Infect Dis. 2004 Mar 1;189(5):852-61. doi: 10.1086/380133. Epub 2004 Feb 6. J Infect Dis. 2004. PMID: 14976603
-
An anti-idiotype vaccine for AIDS based on the HIV receptor.Ann Ist Super Sanita. 1991;27(1):27-31. Ann Ist Super Sanita. 1991. PMID: 1683525 Review.
-
Interactions of HIV-1 with antigen-presenting cells.Immunol Cell Biol. 1999 Aug;77(4):289-303. doi: 10.1046/j.1440-1711.1999.00833.x. Immunol Cell Biol. 1999. PMID: 10457195 Review.
Cited by
-
Reconstruction of a pathway of antigen processing and class II MHC peptide capture.EMBO J. 2007 Apr 18;26(8):2137-47. doi: 10.1038/sj.emboj.7601660. Epub 2007 Mar 29. EMBO J. 2007. PMID: 17396153 Free PMC article.
-
In vivo alteration of humoral responses to HIV-1 envelope glycoprotein gp120 by antibodies to the CD4-binding site of gp120.Virology. 2008 Mar 15;372(2):409-20. doi: 10.1016/j.virol.2007.10.044. Epub 2007 Dec 4. Virology. 2008. PMID: 18054978 Free PMC article.
-
Antibodies to the CD4-binding site of HIV-1 gp120 suppress gp120-specific CD4 T cell response while enhancing antibody response.Infect Agent Cancer. 2008 Jul 18;3:11. doi: 10.1186/1750-9378-3-11. Infect Agent Cancer. 2008. PMID: 18638381 Free PMC article.
-
The use of immune complex vaccines to enhance antibody responses against neutralizing epitopes on HIV-1 envelope gp120.Vaccine. 2009 Dec 11;28(2):352-60. doi: 10.1016/j.vaccine.2009.10.040. Epub 2009 Oct 29. Vaccine. 2009. PMID: 19879224 Free PMC article.
-
The antigenic determinants on HIV p24 for CD4+ T cell inhibiting antibodies as determined by limited proteolysis, chemical modification, and mass spectrometry.J Am Soc Mass Spectrom. 2006 Nov;17(11):1560-1569. doi: 10.1016/j.jasms.2006.06.011. Epub 2006 Jul 27. J Am Soc Mass Spectrom. 2006. PMID: 16875837
References
-
- Botarelli, P., B. A. Houlden, N. L. Haigwood, C. Servis, D. Montagna, and S. Abrignani. 1991. N-glycosylation of HIV-gp120 may constrain recognition by T lymphocytes. J. Immunol. 147:3128-3132. - PubMed
-
- Chien, P. C., Jr., D. Chen, P. Chen, M. Tuen, S. Cohen, S. A. Migueles, M. Connors, E. Rosenberg, U. Malhotra, C. Gonzalez, and C. E. Hioe. 2004. Human immunodeficiency virus type 1-infected patients with envelope-specific lymphoproliferation and long-term nonprogressors lack antibodies suppressing gp120 antigen presentation. J. Infect. Dis. 189:852-861. - PubMed
-
- Chien, P. C., Jr., S. Cohen, C. Kleeberger, J. Giorgi, J. Phair, S. Zolla-Pazner, and C. E. Hioe. 2002. High levels of antibodies to the CD4 binding domain of human immunodeficiency virus type 1 glycoprotein 120 are associated with faster disease progression. J. Infect. Dis. 186:205-213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

