New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a cercopithecus monkey virus clade
- PMID: 15220449
- PMCID: PMC434087
- DOI: 10.1128/JVI.78.14.7748-7762.2004
New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a cercopithecus monkey virus clade
Abstract
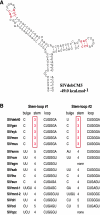
Nearly complete sequences of simian immunodeficiency viruses (SIVs) infecting 18 different nonhuman primate species in sub-Saharan Africa have now been reported; yet, our understanding of the origins, evolutionary history, and geographic distribution of these viruses still remains fragmentary. Here, we report the molecular characterization of a lentivirus (SIVdeb) naturally infecting De Brazza's monkeys (Cercopithecus neglectus). Complete SIVdeb genomes (9,158 and 9227 bp in length) were amplified from uncultured blood mononuclear cell DNA of two wild-caught De Brazza's monkeys from Cameroon. In addition, partial pol sequences (650 bp) were amplified from four offspring of De Brazza's monkeys originally caught in the wild in Uganda. Full-length (9068 bp) and partial pol (650 bp) SIVsyk sequences were also amplified from Sykes's monkeys (Cercopithecus albogularis) from Kenya. Analysis of these sequences identified a new SIV clade (SIVdeb), which differed from previously characterized SIVs at 40 to 50% of sites in Pol protein sequences. The viruses most closely related to SIVdeb were SIVsyk and members of the SIVgsn/SIVmus/SIVmon group of viruses infecting greater spot-nosed monkeys (Cercopithecus nictitans), mustached monkeys (Cercopithecus cephus), and mona monkeys (Cercopithecus mona), respectively. In phylogenetic trees of concatenated protein sequences, SIVdeb, SIVsyk, and SIVgsn/SIVmus/SIVmon clustered together, and this relationship was highly significant in all major coding regions. Members of this virus group also shared the same number of cysteine residues in their extracellular envelope glycoprotein and a high-affinity AIP1 binding site (YPD/SL) in their p6 Gag protein, as well as a unique transactivation response element in their viral long terminal repeat; however, SIVdeb and SIVsyk, unlike SIVgsn, SIVmon, and SIVmus, did not encode a vpu gene. These data indicate that De Brazza's monkeys are naturally infected with SIVdeb, that this infection is prevalent in different areas of the species' habitat, and that geographically diverse SIVdeb strains cluster in a single virus group. The consistent clustering of SIVdeb with SIVsyk and the SIVmon/SIVmus/SIVgsn group also suggests that these viruses have evolved from a common ancestor that likely infected a Cercopithecus host in the distant past. The vpu gene appears to have been acquired by a subset of these Cercopithecus viruses after the divergence of SIVdeb and SIVsyk.
Figures






References
-
- Bailes, E., R. R. Chaudhuri, M. L. Santiago, F. Bibollet-Ruche, B. H. Hahn, and P. M. Sharp. 2002. The evolution of primate lentiviruses and the origins of AIDS, p. 65-96. In T. A. Leitner (ed.), The molecular epidemiology of human viruses. Kluwer Academic Publishers, Boston, Mass.
-
- Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713.. - PubMed
-
- Beer, B. E., E. Bailes, G. Dapolito, B. J. Campbell, R. M. Goeken, M. K. Axthelm, P. D. Markham, J. Bernard, D. Zagury, G. Franchini, P. M. Sharp, and V. M. Hirsch. 2000. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J. Virol. 74:3892-3898. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

