Activity-based probes for the proteomic profiling of metalloproteases
- PMID: 15220480
- PMCID: PMC454150
- DOI: 10.1073/pnas.0402784101
Activity-based probes for the proteomic profiling of metalloproteases
Abstract
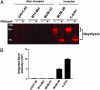
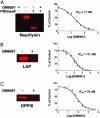
Metalloproteases (MPs) are a large and diverse class of enzymes implicated in numerous physiological and pathological processes, including tissue remodeling, peptide hormone processing, and cancer. MPs are tightly regulated by multiple posttranslational mechanisms in vivo, hindering their functional analysis by conventional genomic and proteomic methods. Here we describe a general strategy for creating activity-based proteomic probes for MPs by coupling a zinc-chelating hydroxamate to a benzophenone photocrosslinker, which promote selective binding and modification of MP active sites, respectively. These probes labeled active MPs but not their zymogen or inhibitor-bound counterparts and were used to identify members of this enzyme class up-regulated in invasive cancer cells and to evaluate the selectivity of MP inhibitors in whole proteomes. Interestingly, the matrix metalloproteinase inhibitor GM6001 (ilomastat), which is currently in clinical development, was found to also target the neprilysin, aminopeptidase, and dipeptidylpeptidase clans of MPs. These results demonstrate that MPs can display overlapping inhibitor sensitivities despite lacking sequence homology and stress the need to evaluate MP inhibitors broadly across this enzyme class to develop agents with suitable target selectivities in vivo. Activity-based profiling offers a powerful means for conducting such screens, as this approach can be carried out directly in whole proteomes, thereby facilitating the discovery of disease-associated MPs concurrently with inhibitors that selectively target these proteins.
Figures



References
-
- Patterson, S. D. & Aebersold, R. (2003) Nat. Genet. 33, 311-323. - PubMed
-
- Adam, G. C., Sorensen, E. J. & Cravatt, B. F. (2002) Mol. Cell. Proteomics 1, 781-790. - PubMed
-
- Michnick, S. W. (2004) Drug Discovery Today 9, 262-267. - PubMed
-
- Patton, W. F., Schulenberg, B. & Steinberg, T. H. (2002) Curr. Opin. Biotechnol. 13, 321-328. - PubMed
-
- Gygi, S. P., Rist, B., Gerber, S. A., Turecek, F., Gelb, M. H. & Aebersold, R. (1999) Nat. Biotechnol. 17, 994-999. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

