Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs
- PMID: 15226410
- PMCID: PMC443529
- DOI: 10.1093/nar/gkh653
Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs
Erratum in
- Nucleic Acids Res. 2004 Aug 17;32(14):4420
Abstract
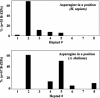
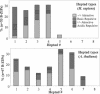
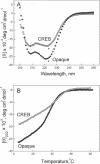
Basic region-leucine zipper (B-ZIP) proteins are a class of dimeric sequence-specific DNA-binding proteins unique to eukaryotes. We have identified 67 B-ZIP proteins in the Arabidopsis thaliana genome. No A.thaliana B-ZIP domains are homologous with any Homo sapiens B-ZIP domains. Here, we predict the dimerization specificity properties of the 67 B-ZIP proteins in the A.thaliana genome based on three structural properties of the dimeric alpha-helical leucine zipper coiled coil structure: (i) length of the leucine zipper, (ii) placement of asparagine or a charged amino acid in the hydrophobic interface and (iii) presence of interhelical electrostatic interactions. Many A.thaliana B-ZIP leucine zippers are predicted to be eight or more heptads in length, in contrast to the four or five heptads typically found in H.sapiens, a prediction experimentally verified by circular dichroism analysis. Asparagine in the a position of the coiled coil is typically observed in the second heptad in H.sapiens. In A.thaliana, asparagine is abundant in the a position of both the second and fifth heptads. The particular placement of asparagine in the a position helps define 14 families of homodimerizing B-ZIP proteins in A.thaliana, in contrast to the six families found in H.sapiens. The repulsive interhelical electrostatic interactions that are used to specify heterodimerizing B-ZIP proteins in H.sapiens are not present in A.thaliana. Instead, we predict that plant leucine zippers rely on charged amino acids in the a position to drive heterodimerization. It appears that A.thaliana define many families of homodimerizing B-ZIP proteins by having long leucine zippers with asparagine judiciously placed in the a position of different heptads.
Figures



Similar articles
-
B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners.Genome Res. 2002 Aug;12(8):1190-200. doi: 10.1101/gr.67902. Genome Res. 2002. PMID: 12176927 Free PMC article.
-
Deciphering B-ZIP transcription factor interactions in vitro and in vivo.Biochim Biophys Acta. 2006 Jan-Feb;1759(1-2):4-12. doi: 10.1016/j.bbaexp.2005.12.005. Epub 2006 Jan 31. Biochim Biophys Acta. 2006. PMID: 16580748 Review.
-
Experimental identification of homodimerizing B-ZIP families in Homo sapiens.J Struct Biol. 2006 Aug;155(2):130-9. doi: 10.1016/j.jsb.2006.02.018. Epub 2006 May 6. J Struct Biol. 2006. PMID: 16725346
-
Preferential heterodimeric parallel coiled-coil formation by synthetic Max and c-Myc leucine zippers: a description of putative electrostatic interactions responsible for the specificity of heterodimerization.J Mol Biol. 1995 Dec 1;254(3):505-20. doi: 10.1006/jmbi.1995.0634. J Mol Biol. 1995. PMID: 7490766
-
Interactions of coiled coils in transcription factors: where is the specificity?Curr Opin Genet Dev. 1993 Apr;3(2):278-85. doi: 10.1016/0959-437x(93)90035-n. Curr Opin Genet Dev. 1993. PMID: 8504253 Review.
Cited by
-
A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes.Plant Cell. 2007 Nov;19(11):3379-90. doi: 10.1105/tpc.107.055772. Epub 2007 Nov 30. Plant Cell. 2007. PMID: 18055602 Free PMC article.
-
GmZPR3d Interacts with GmHD-ZIP III Proteins and Regulates Soybean Root and Nodule Vascular Development.Int J Mol Sci. 2019 Feb 14;20(4):827. doi: 10.3390/ijms20040827. Int J Mol Sci. 2019. PMID: 30769886 Free PMC article.
-
Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis.Mol Biotechnol. 2015 Jan;57(1):12-26. doi: 10.1007/s12033-014-9797-2. Mol Biotechnol. 2015. PMID: 25173685
-
bZIPs and WRKYs: two large transcription factor families executing two different functional strategies.Front Plant Sci. 2014 Apr 30;5:169. doi: 10.3389/fpls.2014.00169. eCollection 2014. Front Plant Sci. 2014. PMID: 24817872 Free PMC article. Review.
-
A post-gene silencing bioinformatics protocol for plant-defence gene validation and underlying process identification: case study of the Arabidopsis thaliana NPR1.BMC Plant Biol. 2017 Nov 23;17(1):218. doi: 10.1186/s12870-017-1151-y. BMC Plant Biol. 2017. PMID: 29169324 Free PMC article.
References
-
- Ellenberger T.E., Brandl,C.J., Struhl,K. and Harrison,S.C. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell, 71, 1223–1237. - PubMed
-
- Vinson C.R., Sigler,P.B. and McKnight,S.L. (1989) Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science, 246, 911–916. - PubMed
-
- Newman J.R. and Keating,A.E. (2003) Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science, 300, 2097–2101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

